Key Points
-
Auxins form a class of small indolic plant growth regulators that was initially identified as the motile stimulus that causes plants to bend towards the light. Auxins have profound effects on many aspects of plant development — they affect cell division, elongation and differentiation — but their modes of action are complicated.
-
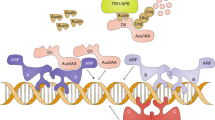
The most well-characterized auxin signalling events are mediated by auxin-responsive elements (AREs) in the promoters of auxin primary-response genes. ARE-mediated transcription is initiated through the action of auxin response factors (ARFs) to AREs. ARE-mediated transcription is inhibited by the binding of Aux/IAA proteins to ARFs.
-
Auxin functions by stimulating the ubiquitin-mediated proteolysis of Aux/IAAs. Aux/IAAs are marked for proteolysis by the action of the SCFTIR1 E3 ubiquitin ligase. The association between TIR1 and Aux/IAAs is enhanced by the direct binding of auxin to TIR1. TIR1 is one of a family of four similar F-box-domain-containing proteins, which all mediate an auxin response and represent the first confirmed auxin receptors.
-
These F-box proteins probably do not represent the only mechanism of auxin perception. Other auxin-binding proteins such as AUXIN-BINDING PROTEIN-1 (ABP1) also confer auxin sensitivity and might also represent auxin receptors. These other pathways are likely to be extremely rapid, and certain auxin responses are seen almost instantaneously.
-
The phytohormone that best fits the classical definition of a hormone is auxin as it is transported between its site of synthesis and site of action. This transport is mediated by several transporters and signalling events that are separate from ARE-mediated gene transcription.
-
Although not traditionally considered together, it is becoming increasingly clear that auxin signalling and auxin transport are linked. The involvement of an increasing number of proteins that were previously considered as components of signalling networks that are also involved in auxin transport is currently opening a new chapter in auxin biology.
Abstract
Hormones have been at the centre of plant physiology research for more than a century. Research into plant hormones (phytohormones) has at times been considered as a rather vague subject, but the systematic application of genetic and molecular techniques has led to key insights that have revitalized the field. In this review, we will focus on the plant hormone auxin and its action. We will highlight recent mutagenesis and molecular studies, which have delineated the pathways of auxin transport, perception and signal transduction, and which together define the roles of auxin in controlling growth and patterning.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sachs, J. Vorlesungen über Pflanzenphysiologie (Engelmann, Leipzig, 1887) (in German).
Darwin, F. & Darwin, C. The Power of Movement in Plants (John Murray, London, 1880).
Cholodny, N. Beiträge zur Analyse der geotropischen Reaktion. Jahrbuch Wiss. Bot. 65, 447–459 (1926) (in German).
Went, F. On growth accelerating substances in the coleoptile of Avena sativa. Proc. K. Akad. Wetensch. Amsterdam 30, 10–19 (1926).
Folkes, L. K. & Wardman, P. Oxidative activation of indole-3-acetic acids to cytotoxic species — a potential new role for plant auxins in cancer therapy. Biochem. Pharmacol. 61, 129–136 (2001).
Folkes, L. K. & Wardman, P. Enhancing the efficacy of photodynamic cancer therapy by radicals from plant auxin (indole-3-acetic acid). Cancer Res. 63, 776–779 (2003).
Theologis, A. & Ray, P. M. Early auxin-regulated polyadenylylated mRNA sequences in pea stem tissue. Proc. Natl Acad. Sci. USA 79, 418–421 (1982).
Theologis, A., Huynh, T. V. & Davis, R. W. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 183, 53–68 (1985).
Key, J. L., Barnett, N. M. & Lin, C. Y. RNA and protein biosynthesis and the regulation of cell elongation by auxin. Ann. NY Acad. Sci. 144, 49–62 (1967).
Abel, S. & Theologis, A. Early genes and auxin action. Plant Physiol. 111, 9–17 (1996).
Leyser, H. M. et al. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164 (1993).
del Pozo, J. C. & Estelle, M. Function of the ubiquitin–proteosome pathway in auxin response. Trends Plant Sci. 4, 107–112 (1999).
del Pozo, J. C., Timpte, C., Tan, S., Callis, J. & Estelle, M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763 (1998).
McClure, B. A. & Guilfoyle, T. J. Characterisation of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 9, 611–623 (1987).
Hagen, G. & Guilfoyle, T. J. Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203 (1985).
Remington, D. L., Vision, T. J., Guilfoyle, T. J. & Reed, J. W. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752 (2004).
Okushima, Y. et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463 (2005).
Overvoorde, P. J. et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17, 3282–3300 (2005).
Beh, C. T., Cool, L., Phillips, J. & Rine, J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157, 1117–1140 (2001).
Ulmasov, T., Murfett, J., Hagen, G. & Guilfoyle, T. J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin responsive elements. Plant Cell 9, 1963–1971 (1997). The authors created a highly active synthetic ARE (known as DR5) that has become the standard tool for studying auxin-responsive transcription. This paper shows that overexpression of Aux/IAA proteins results in the specific repression of the DR5 reporter gene.
Ballas, N., Wong, L. M. & Theologis, A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 233, 580–96 (1993).
Ulmasov, T., Hagen, G. & Guilfoyle, T. J. Activation and repression of transription by auxin-response factors. Proc. Natl Acad. Sci. USA 96, 5844–5849 (1999).
Ulmasov, T., Hagen, G. & Guilfoyle, T. J. ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 (1997).
Ouellet, F., Overvoorde, P. J. & Theologis, A. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841 (2001).
Leyser, H. M., Pickett, F. B., Dharmasiri, S. & Estelle, M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413 (1996).
Worley, C. K. et al. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562 (2000).
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001).
Ramos, J. A., Zenser, N., Leyser, O. & Callis, J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360 (2001).
Dharmasiri, N. & Estelle, M. Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308 (2004).
Bernard, F. & Andre, B. Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496, 81–85 (2001).
Karin, M. & Ben-Neriah, Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18, 621–663 (2000).
Sullivan, J. A., Shirasu, K. & Deng, X. W. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nature Rev. Genet. 4, 948–958 (2003).
Schwechheimer, C. et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382 (2001).
Chuang, H. W., Zhang, W. & Gray, W. M. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. Plant Cell 16, 1883–1897 (2004).
Xiao, W. Y. & Jang, J. C. F-box proteins in Arabidopsis. Trends Plant Sci. 5, 454–457 (2000).
Callis, J. & Vierstra, R. D. Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386 (2000).
Gray, W. M. & Estelle, M. Function of the ubiquitin–proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138 (2000).
Leyser, O. Auxin signalling: the beginning, the middle and the end. Curr. Opin. Plant Biol. 4, 382–386 (2001).
Estelle, M. Proteases and cellular regulation in plants. Curr. Opin. Plant Biol. 4, 254–260 (2001).
Ward, S. P. & Estelle, M. Auxin signaling involves regulated protein degradation by the ubiquitin–proteasome pathway. J. Plant Growth Regul. 20, 265–273 (2001).
Kepinski, S. & Leyser, O. Ubiquitination and auxin signaling: a degrading story. Plant Cell 14, S81–S95 (2002).
Vierstra, R. D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142 (2003).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005).
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005). References 43 and 44 both show that TIR1 is an auxin receptor that mediates Aux/IAA degradation and auxin-regulated transcription.
Dharmasiri, N., Dharmasiri, S., Jones, A. M. & Estelle, M. Auxin action in a cell-free system. Curr. Biol. 13, 1418–1422 (2003).
Kepinski, S. & Leyser, O. Auxin-induced SCFTIR1–Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl Acad. Sci. USA 101, 12381–12386 (2004).
Risseeuw, E. P. et al. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34, 753–767 (2003).
Gagne, J. M. et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl Acad. Sci. USA 101, 6803–6808 (2004).
Smalle, J. & Vierstra, R. D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590 (2004).
Dharmasiri, N. et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005).
del Pozo, J. C. & Estelle, M. F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol. Biol. 44, 123–128 (2000).
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998).
Sasaki, A. et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898 (2003).
Kim, J., Harter, K. & Theologis, A. Protein–protein interactions among the Aux/IAA proteins. Proc. Natl Acad. Sci. USA 94, 11786–11791 (1997).
Tiwari, S. B., Hagen, G. & Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543 (2003).
Hardtke, C. S. et al. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100 (2004).
Ellis, C. M. et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574 (2005).
Okushima, Y., Mitina, I., Quach, H. L. & Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46 (2005).
Wilmoth, J. C. et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130 (2005).
Weijers, D. et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24, 1874–1885 (2005).
Reinhardt, D. et al. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260 (2003).
Benkova, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Aida, M. et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Venis, M. A., Thomas, E. W., Barbier, B. H., Ephritikhine, G. & Guern, J. Impermeant auxin analogues have auxin activity. Planta 182, 232–235 (1990).
Felle, H., Peters, W. & Palme, K. The electrical response of maize to auxins. Biochim. Biophys. Acta 1064, 199–204 (1991).
Frias, I. et al. A major isoform of the maize plasma membrane H+-ATPase: characterization and induction by auxin in coleoptiles. Plant Cell 8, 1533–1544 (1996).
Hager, A., Debus, G., Edel, H. G., Stransky, H. & Serrano, R. Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H+-ATPase. Planta 185, 527–537 (1991).
Rück, A., Palme, K., Venis, M. A., Napier, R. M. & Felle, R. H. Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 4, 41–46 (1993).
Senn, A. P. & Goldsmith, M. H. M. Regulation of electrogenic proton pumping by auxin and fusicoccin as related to the growth of Avena coleoptiles. Plant Physiol. 88, 131–138 (1988).
Blatt, M. R. & Thiel, G. K+ channels of stomatal guard cells: bimodal control of the K+ inward-rectifier evoked by auxin. Plant J. 5, 55–68 (1994).
Marten, I., Lohse, G. & Hedrich, R. Plant growth hormones control voltage-dependent activity of anion channels in plasma membrane of guard cells. Nature 353, 758–762 (1991).
Philippar, K. et al. Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc. Natl Acad. Sci. USA 96, 12186–12191 (1999).
Zimmermann, S., Thomine, S., Guern, J. & Barbier-Brygoo, H. An anion current at the plasma membrane of tobacco protoplasts shows ATP-dependent voltage regulation and is modulated by auxin. Plant J. 6, 707–716 (1994).
Evans, M. L. The action of auxin on plant cell elongation. Crit. Rev. Plant Sci. 2, 317–365 (1985).
Palme, K. From binding proteins to hormone receptors? J. Plant Growth Regul. 12, 171–178 (1994).
Napier, R. M. Towards an understanding of ABP1. J. Exp. Bot. 46, 1787–1795 (1995).
Hesse, T. et al. Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J. 8, 2453–2461 (1989).
Woo, E.-J. et al. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 21, 2877–2885 (2002).
Dunwell, J. M., Khuri, S. & Gane, P. J. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179 (2000).
Chen, J. G., Shimomura, S., Sitbon, F., Sandberg, G. & Jones, A. M. The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J. 28, 607–617 (2001).
Gehring, C. A., McConchie, R. M., Venis, M. A. & Parish, R. W. Auxin-binding-protein antibodies and peptides influence stomatal opening and alter cytoplasmic pH. Planta 205, 581–586 (1998).
Leblanc, N., Perrot-Rechenmann, C. & Barbier-Brygoo, H. The auxin-binding protein Nt-ERabp1 alone activates an auxin-like transduction pathway. FEBS Lett. 449, 57–60 (1999).
Jones, A. M. et al. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282, 1114–1117 (1998).
Steffens, B. et al. The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J. 27, 591–599 (2001).
Chen, J. G., Ullah, H., Young, J. C., Sussman, M. R. & Jones, A. M. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15, 902–911 (2001). After many years of investigation, the importance of ABP1 for both cell elongation and cell division was clearly shown. Crucially, the loss-of-function abp1 mutant has an embryonic-lethal phenotype.
Laskowski, M. J. et al. FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol. 128, 578–590 (2002).
Foreman, J. et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446 (2003).
Schopfer, P. Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J. 28, 679–688 (2001).
Hancock, J. T., Desikan, R. & Neill, S. J. Role of reactive oxygen species in cell signalling pathways. Biochem. Soc. Trans. 29, 345–350 (2001).
Vanneste, S. et al. Auxin regulation of cell cycle and its role during lateral root initiation. Physiol. Plantarum 123, 139–146 (2005).
Richard, C., Lescot, M., Inze, D. & De Veylder, L. Effect of auxin, cytokinin, and sucrose on cell cycle gene expression in Arabidopsis thaliana cell suspension cultures. Plant Cell Tissue Organ Cult. 69, 167–176 (2002).
del Pozo, J. C., Boniotti, M. B. & Gutierrez, C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14, 3057–3071 (2002).
Blilou, I. et al. The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16, 2566–2575 (2002).
Bartel, B. Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 49–64 (1997).
Normanly, J. Auxin metabolism. Physiol. Plantarum 100, 431–442 (1997).
Ljung, K., Bhalerao, R. P. & Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474 (2001).
Ljung, K. et al. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 49, 249–272 (2002).
Ljung, K. et al. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104 (2005). Until this paper, it was widely believed that auxin was exclusively synthesized in the shoot.
Woodward, A. W. & Bartel, B. Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 (2005).
Noh, B., Murphy, A. S. & Spalding, E. P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13, 2441–2454 (2001).
Geisler, M. et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14, 4238–4249 (2003).
Vicente-Agullo, F. et al. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 40, 523–535 (2004).
Li, J. S. et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310, 121–125 (2005).
Lin, R. C. & Wang, H. Y. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 138, 949–964 (2005).
Friml, J., Wisniewska, J., Benkova, E., Mendgen, K. & Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002). PIN3 can relocalize in response to gravity. This is the first report that PIN polarity can change in response to an environmental stimulus.
Friml, J. et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673 (2002).
Gälweiler, L. et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998). This paper shows that proteins can be localized polarly in plants. A correlation with the expected direction of auxin transport indicates that PIN is an auxin efflux carrier.
Müller, A. et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911 (1998).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Bennett, M. J. et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–959 (1996). The authors present a detailed characterization of the auxin influx carrier and its function in the gravitropic response.
Paponov, I. A., Teale, W. D., Trebar, M., Blilou, K. & Palme, K. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10, 170–177 (2005).
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J. & Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684 (1991).
Ottenschläger, I. et al. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl Acad. Sci. USA 100, 2987–2991 (2003).
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005).
Swarup, R. et al. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648–2653 (2001).
Marchant, A. et al. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14, 589–597 (2002).
Marchant, A. et al. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18, 2066–2073 (1999).
Yang, Y., Hammes, U. Z., Taylor, C. G., Schachtman, D. P. & Nielsen, E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16, 1123–1127 (2006).
Dharmasiri, S. et al. AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312, 1218–1220 (2006).
Wisniewska, J. et al. Polar PIN localization directs auxin flow in plants. Science 312, 883–883 (2006).
Petrasek, J. et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 (2006).
Geldner, N., Friml, J., Stierhof, Y. D., Jürgens, G. & Palme, K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428 (2001). Shows that the seemingly static localization of PIN1 is maintained by rapid cycling between the plasma membrane and endosomal compartments.
Paciorek, T. et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256 (2005).
Noh, B., Bandyopadhyay, A., Peer, W. A., Spalding, E. P. & Murphy, A. S. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423, 999–1002 (2003).
Vieten, A. et al. Functional redundancy of PIN proteins is accompanied by auxin dependent cross-regulation of PIN expression. Development 132, 4521–4531 (2005).
Xu, J. et al. A molecular framework for plant regeneration. Science 311, 385–388 (2006).
Weijers, D. et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270 (2006).
Steinmann, T. et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318 (1999).
Geldner, N. et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 (2003). The authors engineered a brefeldin A (BFA)-resistant version of GNOM, and, in plants that harbour this fully functional GNOM variant, PIN1 localization and auxin transport are no longer sensitive to BFA. It was therefore suggested that ARF GEFs regulate specific endosomal trafficking pathways.
Geldner, N. et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131, 389–400 (2004).
Garbers, C. et al. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15, 2115–2124 (1996).
Rashotte, A. M., DeLong, A. & Muday, G. K. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13, 1683–1697 (2001).
Shin, H. et al. Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J. 42, 188–200 (2005).
Furutani, M. et al. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030 (2004).
Benjamins, R., Quint, A., Weijers, D., Hooykaas, P. & Offringa, R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067 (2001).
Christensen, S. K., Dagenais, N., Chory, J. & Weigel, D. Regulation of auxin response by the protein kinase PINOID. Cell 100, 469–478 (2000).
Friml, J. et al. A PINOID-dependent binary switch in apical–basal PIN polar targeting directs auxin efflux. Science 306, 862–865 (2004). It was demonstrated that the kinase PINOID controls PIN polarity and is necessary for correct cellular patterning.
Benjamins, R., Ampudia, C. S. G., Hooykaas, P. J. J. & Offringa, R. PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 132, 1623–1630 (2003).
Zegzouti, H., Anthony, R. G., Jahchan, N., Bogre, L. & Christensen, S. K. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 6404–6409 (2006).
Beltran-Pena, E., Aguilar, R., Ortiz-Lopez, A., Dinkova, T. D. & de Jimenez, E. S. Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol. Plantarum 115, 291–297 (2002).
Kovtun, Y., Chiu, W. L., Tena, G. & Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl Acad. Sci. USA 97, 2940–2945 (2000).
Mockaitis, K. & Howell, S. H. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796 (2000).
Dai, Y. et al. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18, 308–320 (2006).
Hössel, D., Schmeiser, C. & Hertel, R. Specificity patterns indicate that auxin exporters and receptors are the same proteins. Plant Biol. 7, 41–48 (2005).
Tao, L. Z., Cheung, A. Y. & Wu, H. M. Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14, 2745–2760 (2002).
Tao, L. Z., Cheung, A. Y., Nibau, C. & Wu, H. M. RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell 17, 2369–2383 (2005).
Rhoades, M. W. et al. Prediction of plant microRNA targets. Cell 110, 513–520 (2002).
Wang, J. W. et al. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216 (2005).
Fahlgren, N. et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16, 939–944 (2006).
Guo, H. S., Xie, Q., Fei, J. F. & Chua, N. H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386 (2005).
Xie, Q. et al. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170 (2002).
Navarro, L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006).
Fleming, A. Formation of primordia and phyllotaxy. Curr. Opin. Plant Biol. 8, 53–58 (2005).
Aloni, R. in Plant Hormones — Biosynthesis, Signal Transduction, Action! (ed. Davis, P. J.) 471–492 (Kluwer, Dordrecht, 2004).
Teale, W. D., Paponov, I. A., Ditengou, F. & Palme, K. Auxin and the developing root of Arabidopsis thaliana. Physiol. Plantarum 123, 130–138 (2005).
Ruegger, M. et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207 (1998).
Hertel, R., Evans, M. R., Leopold, A. C. & Sell, H. M. The specificity of the auxin transport system. Planta 85, 238–249 (1969).
Acknowledgements
We thank W. Hartung, R. Hertel, S. Kepinski, J. McWilliams and P. Schopfer for critical reading of the manuscript. Our work was supported by the Deutsche Forschungsgemeinschaft (DFG), the Bundesministerium für Bildung und Forschung, Fonds der chemischen Industrie, the European Union and the Landesstiftung Baden-Württemberg GmbH. K.P. is particularly grateful for having had the opportunity to contribute to the DFG network's Molecular Mechanisms of Phytohormone Action.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Regulon
-
A collection of separate genes, the expression of which is controlled as a unit by a specific signalling compound or factor.
- Degron
-
A protein element, usually a sequence motif, that targets the protein for proteolytic degradation.
- SCFTIR1 E3 ubiquitin ligase
-
A multisubunit ubiquitin ligase that consists of SKP1, CUL1 and an F-box protein (TIR1 in this case) that confers substrate specificity, as well as a RING protein that is also known as HRT1, RBX1 or ROC1.
- F-box protein
-
A component of the machinery for the ubiquitin-dependent degradation of proteins. F-box proteins recognize specific substrates and, with the help of other subunits of the E3 ubiquitin ligase, deliver them to the E2 ubiquitin-conjugating enzyme.
- 26S proteasome
-
A protein complex that is responsible for degrading intracellular proteins that have been tagged for destruction by the addition of ubiquitin.
- COP9 signalosome
-
An eight-subunit protein complex that regulates protein ubiquitylation and turnover in a range of developmental and physiological contexts. Extensively characterized in plants but fundamental to all eukaryotes, this complex post-translationally modifies the cullin subunit of E3-ubiquitin ligases by cleaving off the covalently coupled peptide, Nedd8/RUB1.
- T-DNA mutation
-
A mutation that is the result of the integration of DNA from Agrobacterium tumefaciens into plant genomes. The insertion is random and might therefore disrupt genes, causing a mutation at the insertion point.
- Cupin proteins
-
A diverse family of plant proteins, all of which contain at least one double-stranded α-helix or jelly-roll structural motif. This motif is also present in all structurally characterized 2-oxoglutarate-dependent oxygenases, including the hypoxia-inducible factor (HIF) hydroxylases, and is characteristic of the Jumonji transcription factors.
- Root cap
-
The layers of protective cells that cover the tip of the growing root.
- Expansins
-
A class of proteins that are able to catalyse the loosening of the cell wall, enabling cell expansion.
- Extensins
-
Proline-rich proteins that connect the cell wall and the plasma membrane.
- Meristem
-
A zone (for example, the apex of the shoot) that contains undifferentiated cells that continue to divide, providing cells for further growth and differentiation.
- Phloem
-
Vascular tissue that carries organic nutrients as well as information molecules such as hormones throughout the plant.
- Xylem
-
Vascular tissue that delivers water and mineral nutrients, which are taken up by the root system, to aerial organs. It also provides mechanical support.
- Agravitropic
-
When a plant is unable to either perceive or respond to gravity. Typically, the roots of agravitropic plants grow in all directions.
- Guanine nucleotide exchange factor
-
A protein that facilitates the exchange of GDP (guanine diphosphate) for GTP (guanine triphosphate) in the nucleotide-binding pocket of a GTP-binding protein.
- GTPase-activating protein
-
(GAP). A protein that stimulates the intrinsic ability of a GTPase to hydrolyse GTP to GDP. Therefore, GAPs negatively regulate GTPases by converting them from active (GTP-bound) to inactive (GDP-bound).
Rights and permissions
About this article
Cite this article
Teale, W., Paponov, I. & Palme, K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7, 847–859 (2006). https://doi.org/10.1038/nrm2020
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2020
This article is cited by
-
Genome-wide identification and expression characterization of the GH3 gene family of tea plant (Camellia sinensis)
BMC Genomics (2024)
-
Analysis of the global transcriptome and miRNAome associated with seed dormancy during seed maturation in rice (Oryza sativa L. cv. Nipponbare)
BMC Plant Biology (2024)
-
BRIP1 and BRIP2 maintain root meristem by affecting auxin-mediated regulation
Planta (2024)
-
Aldoximes: compounds at the crossroads of multiple metabolic pathways in plant
Phytochemistry Reviews (2024)
-
Transcriptomics reveals a core transcriptional network of K-type cytoplasmic male sterility microspore abortion in wheat (Triticum aestivum L.)
BMC Plant Biology (2023)