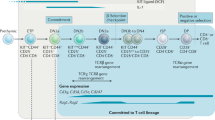
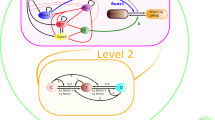
Key Points
-
To gain a more detailed understanding of T cell differentiation, methods are required with which the fate of T cells can be followed at the single-cell level.
-
Three different experimental strategies have been developed that can be used to follow cell fate at the single-cell level, namely continuous microscopy, single-cell adoptive transfer and cell marking with unique tags.
-
Single-cell transfer and cellular barcoding, in which individual naive CD8+ T cells are provided with unique genetic tags, have both been used to show that the progeny of single naive T cells can adopt multiple fates.
-
Cellular barcoding has also been used to demonstrate that the magnitude of antigen-specific CD8+ T cell responses following infection is primarily determined by clonal burst size.
-
To couple T cell functional states to subsequent cell-fate decisions, various gene expression reporter mice have been generated. The use of these mouse models has revealed that T cells with an effector phenotype can survive the contraction phase and enter the memory pool.
-
Understanding how prior signalling input regulates subsequent T cell behaviour will require the use of novel technologies that can monitor these signalling events. In many cases, the mapping of these receptor-mediated signals will require the engineering of synthetic signal transduction pathways.
Abstract
The behaviour of T cells is not fixed in the germ line, but is highly adaptable depending on experiences encountered during a T cell's life. To understand how different T cell subsets arise and how prior signalling input regulates subsequent T cell behaviour, approaches are required that couple a given T cell state to signals received by the cell, or by one of its ancestors, at earlier times. Here we describe recently developed technologies that have been used to determine the kinship of different T cell subsets and their prior functional characteristics. Furthermore, we discuss the potential value of new technologies that would allow assessment of T cell migration patterns and prior signalling events.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Blattman, J. N. et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 195, 657–664 (2002).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Obar, J. J., Khanna, K. M. & Lefrancois, L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28, 859–869 (2008).
Butz, E. A. & Bevan, M. J. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity 8, 167–175 (1998).
Murali-Krishna, K. et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8, 177–187 (1998).
Busch, D. H., Pilip, I. M., Vijh, S. & Pamer, E. G. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity 8, 353–362 (1998).
van Stipdonk, M. J., Lemmens, E. E. & Schoenberger, S. P. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nature Immunol. 2, 423–429 (2001).
Wherry, E. J., Puorro, K. A., Porgador, A. & Eisenlohr, L. C. The induction of virus-specific CTL as a function of increasing epitope expression: responses rise steadily until excessively high levels of epitope are attained. J. Immunol. 163, 3735–3745 (1999).
Badovinac, V. P., Porter, B. B. & Harty, J. T. Programmed contraction of CD8+ T cells after infection. Nature Immunol. 3, 619–626 (2002).
Prlic, M., Hernandez-Hoyos, G. & Bevan, M. J. Duration of the initial TCR stimulus controls the magnitude but not functionality of the CD8+ T cell response. J. Exp. Med. 203, 2135–2143 (2006).
Harty, J. T. & Badovinac, V. P. Shaping and reshaping CD8+ T-cell memory. Nature Rev. Immunol. 8, 107–119 (2008).
Williams, M. A. & Bevan, M. J. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25, 171–192 (2007).
Reiner, S. L., Sallusto, F. & Lanzavecchia, A. Division of labor with a workforce of one: challenges in specifying effector and memory T cell fate. Science 317, 622–625 (2007).
Ahmed, R., Bevan, M. J., Reiner, S. L. & Fearon, D. T. The precursors of memory: models and controversies. Nature Rev. Immunol. 9, 662–668 (2009).
Jameson, S. C. & Masopust, D. Diversity in T cell memory: an embarrassment of riches. Immunity 31, 859–871 (2009).
Zhou, L., Chong, M. M. & Littman, D. R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009).
Locksley, R. M. Nine lives: plasticity among T helper cell subsets. J. Exp. Med. 206, 1643–1646 (2009).
King, C. New insights into the differentiation and function of T follicular helper cells. Nature Rev. Immunol. 9, 757–766 (2009).
Weaver, C. T. & Hatton, R. D. Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nature Rev. Immunol. 9, 883–889 (2009).
Greenberg, P. D. & Cheever, M. A. Treatment of disseminated leukemia with cyclophosphamide and immune cells: tumor immunity reflects long-term persistence of tumor-specific donor T cells. J. Immunol. 133, 3401–3407 (1984).
Shen, F. W. et al. Cloning of Ly-5 cDNA. Proc. Natl Acad. Sci. USA 82, 7360–7363 (1985).
Kearney, E. R., Pape, K. A., Loh, D. Y. & Jenkins, M. K. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity 1, 327–339 (1994).
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
Parish, C. R. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 77, 499–508 (1999).
Roberts, A. D., Ely, K. H. & Woodland, D. L. Differential contributions of central and effector memory T cells to recall responses. J. Exp. Med. 202, 123–133 (2005).
Wright, D. E., Wagers, A. J., Gulati, A. P., Johnson, F. L. & Weissman, I. L. Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936 (2001).
Klonowski, K. D. et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity 20, 551–562 (2004).
Merad, M. et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nature Immunol. 3, 1135–1141 (2002).
von Andrian, U. H. & Mempel, T. R. Homing and cellular traffic in lymph nodes. Nature Rev. Immunol. 3, 867–878 (2003).
Miller, M. J., Wei, S. H., Parker, I. & Cahalan, M. D. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296, 1869–1873 (2002).
Stoll, S., Delon, J., Brotz, T. M. & Germain, R. N. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science 296, 1873–1876 (2002).
Bousso, P., Bhakta, N. R., Lewis, R. S. & Robey, E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science 296, 1876–1880 (2002).
Germain, R. N., Miller, M. J., Dustin, M. L. & Nussenzweig, M. C. Dynamic imaging of the immune system: progress, pitfalls and promise. Nature Rev. Immunol. 6, 497–507 (2006).
Cahalan, M. D. & Parker, I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annu. Rev. Immunol. 26, 585–626 (2008).
Bousso, P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nature Rev. Immunol. 8, 675–684 (2008).
Mempel, T. R., Henrickson, S. E. & von Andrian, U. H. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004).
Miller, M. J., Safrina, O., Parker, I. & Cahalan, M. D. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J. Exp. Med. 200, 847–856 (2004).
Hugues, S. et al. Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nature Immunol. 5, 1235–1242 (2004).
Henrickson, S. E. et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nature Immunol. 9, 282–291 (2008).
Bousso, P. & Robey, E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nature Immunol. 4, 579–585 (2003).
Miller, M. J., Hejazi, A. S., Wei, S. H., Cahalan, M. D. & Parker, I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc. Natl Acad. Sci. USA 101, 998–1003 (2004).
Beltman, J. B., Maree, A. F., Lynch, J. N., Miller, M. J. & de Boer, R. J. Lymph node topology dictates T cell migration behavior. J. Exp. Med. 204, 771–780 (2007).
Wu, M. et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554 (2007).
Schroeder, T. Imaging stem-cell-driven regeneration in mammals. Nature 453, 345–351 (2008).
Eilken, H. M., Nishikawa, S. & Schroeder, T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457, 896–900 (2009). This study uses long-term in vitro time-lapse microscopy to show that adherent endothelial cells can directly give rise to non-adherent haematopoietic cells.
Zovein, A. C. et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636 (2008).
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010).
Rieger, M. A., Hoppe, P. S., Smejkal, B. M., Eitelhuber, A. C. & Schroeder, T. Hematopoietic cytokines can instruct lineage choice. Science 325, 217–218 (2009).
Hawkins, E. D., Markham, J. F., McGuinness, L. P. & Hodgkin, P. D. A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proc. Natl Acad. Sci. USA 106, 13457–13462 (2009). This study uses long-term in vitro imaging to reveal that progeny of single founder B cells have markedly synchronized division properties.
Spencer, S. L., Gaudet, S., Albeck, J. G., Burke, J. M. & Sorger, P. K. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009).
Sigal, A. et al. Variability and memory of protein levels in human cells. Nature 444, 643–646 (2006).
Chang, H. H., Hemberg, M., Barahona, M., Ingber, D. E. & Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008).
Feinerman, O., Veiga, J., Dorfman, J. R., Germain, R. N. & Altan-Bonnet, G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science 321, 1081–1084 (2008).
Osawa, M., Hanada, K., Hamada, H. & Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996).
Kiel, M. J. et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005).
Stemberger, C. et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity 27, 985–997 (2007). This study analyses in vivo T cell fate by performing single cell transfer and shows that one naive CD8+ T cell can yield diverse effector and memory T cell subsets.
Kedzierska, K., La Gruta, N. L., Stambas, J., Turner, S. J. & Doherty, P. C. Tracking phenotypically and functionally distinct T cell subsets via T cell repertoire diversity. Mol. Immunol. 45, 607–618 (2008).
Maryanski, J. L., Jongeneel, C. V., Bucher, P., Casanova, J. L. & Walker, P. R. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity 4, 47–55 (1996).
Busch, D. H., Pilip, I. & Pamer, E. G. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J. Exp. Med. 188, 61–70 (1998).
Sourdive, D. J. et al. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J. Exp. Med. 188, 71–82 (1998).
Lin, M. Y. & Welsh, R. M. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J. Exp. Med. 188, 1993–2005 (1998).
McHeyzer-Williams, L. J., Panus, J. F., Mikszta, J. A. & McHeyzer-Williams, M. G. Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs. J. Exp. Med. 189, 1823–1838 (1999).
Blattman, J. N., Sourdive, D. J., Murali-Krishna, K., Ahmed, R. & Altman, J. D. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J. Immunol. 165, 6081–6090 (2000).
Fasso, M. et al. T cell receptor (TCR)-mediated repertoire selection and loss of TCR Vβ diversity during the initiation of a CD4+ T cell response in vivo. J. Exp. Med. 192, 1719–1730 (2000).
Turner, S. J., Diaz, G., Cross, R. & Doherty, P. C. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity 18, 549–559 (2003).
Kedzierska, K., Turner, S. J. & Doherty, P. C. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc. Natl Acad. Sci. USA 101, 4942–4947 (2004).
Malherbe, L., Hausl, C., Teyton, L. & McHeyzer-Williams, M. G. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 21, 669–679 (2004).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Masopust, D., Vezys, V., Marzo, A. L. & Lefrancois, L. Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417 (2001).
Baron, V. et al. The repertoires of circulating human CD8+ central and effector memory T cell subsets are largely distinct. Immunity 18, 193–204 (2003).
Bouneaud, C., Garcia, Z., Kourilsky, P. & Pannetier, C. Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. J. Exp. Med. 201, 579–590 (2005).
Kedzierska, K. et al. Early establishment of diverse T cell receptor profiles for influenza-specific CD8+CD62Lhi memory T cells. Proc. Natl Acad. Sci. USA 103, 9184–9189 (2006).
Wong, J., Mathis, D. & Benoist, C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J. Exp. Med. 204, 2039–2045 (2007).
Jacob, J. & Kelsoe, G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J. Exp. Med. 176, 679–687 (1992).
Schepers, K. et al. Dissecting T cell lineage relationships by cellular barcoding. J. Exp. Med. 205, 2309–2318 (2008).
Gerlach, C. et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J. Exp. Med. 207, 1235–1246 (2010). This study develops barcode tagging of thymocytes and uses it to show that the progeny of single naive CD8+ T cells can take on multiple fates under various infectious challenges.
Masopust, D. et al. Activated and memory CD8 T cells migrate to nonlymphoid tissues regardless of site activation or tissue of origin. J. Immunol. 172, 4875–4882 (2004).
Masopust, D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010).
van Heijst, J. W. et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science 325, 1265–1269 (2009). This paper uses cellular barcoding to show that the magnitude of CD8+ T cell responses is primarily determined by clonal burst size.
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). In this paper, a Cre–lox-based approach is described that leads to the stochastic expression of multiple fluorescent proteins, allowing the identity of individual neurons to be distinguished by expression of 1 out of almost 100 different colour variations.
Hodgkin, P. D., Lee, J. H. & Lyons, A. B. B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 184, 277–281 (1996).
Gett, A. V. & Hodgkin, P. D. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc. Natl Acad. Sci. USA 95, 9488–9493 (1998).
Bird, J. J. et al. Helper T cell differentiation is controlled by the cell cycle. Immunity 9, 229–237 (1998).
Kaech, S. M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Shin, H. & Wherry, E. J. CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19, 408–415 (2007).
O'Shea, J. J. & Paul, W. E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010).
Mohrs, M., Shinkai, K., Mohrs, K. & Locksley, R. M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 15, 303–311 (2001).
Stetson, D. B. et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198, 1069–1076 (2003).
Harrington, L. E., Janowski, K. M., Oliver, J. R., Zajac, A. J. & Weaver, C. T. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452, 356–360 (2008).
Jacob, J. & Baltimore, D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature 399, 593–597 (1999).
Bannard, O., Kraman, M. & Fearon, D. T. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science 323, 505–509 (2009). This paper uses a conditional granzyme B–YFP reporter mouse to show that granzyme B-expressing effector T cells can give rise to functional memory T cells.
O'Connell, R. M., Rao, D. S., Chaudhuri, A. A. & Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nature Rev. Immunol. 10, 111–122 (2010).
Gett, A. V. & Hodgkin, P. D. A cellular calculus for signal integration by T cells. Nature Immunol. 1, 239–244 (2000).
Hawkins, E. D., Turner, M. L., Dowling, M. R., van Gend, C. & Hodgkin, P. D. A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc. Natl Acad. Sci. USA 104, 5032–5037 (2007).
Becker, T. C., Coley, S. M., Wherry, E. J. & Ahmed, R. Bone marrow is a preferred site for homeostatic proliferation of memory CD8 T cells. J. Immunol. 174, 1269–1273 (2005).
Mazo, I. B. et al. Bone marrow is a major reservoir and site of recruitment for central memory CD8+ T cells. Immunity 22, 259–270 (2005).
Tokoyoda, K. et al. Professional memory CD4+ T lymphocytes preferentially reside and rest in the bone marrow. Immunity 30, 721–730 (2009).
Hatta, K., Tsujii, H. & Omura, T. Cell tracking using a photoconvertible fluorescent protein. Nature Protoc. 1, 960–967 (2006).
Tomura, M. et al. Monitoring cellular movement in vivo with photoconvertible fluorescence protein 'Kaede' transgenic mice. Proc. Natl Acad. Sci. USA 105, 10871–10876 (2008). In this paper, the Kaede transgenic mouse model is described, in which photoconversion of a fluorescent protein can be used to track cell migration.
Pryciak, P. M. Designing new cellular signaling pathways. Chem. Biol. 16, 249–254 (2009).
Strickland, D., Moffat, K. & Sosnick, T. R. Light-activated DNA binding in a designed allosteric protein. Proc. Natl Acad. Sci. USA 105, 10709–10714 (2008). This paper provides proof-of-concept for a light-activated transcription factor by fusing a photoactive LOV domain to the E. coli trp repressor and inducing DNA binding by blue light excitation.
Croxford, A. L., Kurschus, F. C. & Waisman, A. Cutting edge: an IL-17F-CreEYFP reporter mouse allows fate mapping of Th17 cells. J. Immunol. 182, 1237–1241 (2009).
Schlenner, S. M. et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010).
Yeh, B. J., Rutigliano, R. J., Deb, A., Bar-Sagi, D. & Lim, W. A. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature 447, 596–600 (2007). This study provides proof-of-concept for a reporter of kinase activity that is based on the combination of a PDZ–peptide autoinhibition moiety and a Rho–GEF reporter domain.
Dueber, J. E., Yeh, B. J., Chak, K. & Lim, W. A. Reprogramming control of an allosteric signaling switch through modular recombination. Science 301, 1904–1908 (2003).
Struhl, G. & Adachi, A. Nuclear access and action of notch in vivo. Cell 93, 649–660 (1998).
Radtke, F., Fasnacht, N. & Robson MacDonald, H. R. Notch signaling in the immune system. Immunity 32, 14–27 (2010).
Howard, P. L., Chia, M. C., Del Rizzo, S., Liu, F. F. & Pawson, T. Redirecting tyrosine kinase signaling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc. Natl Acad. Sci. USA 100, 11267–11272 (2003).
Wu, A. M., Till, J. E., Siminovitch, L. & McCulloch, E. A. A cytological study of the capacity for differentiation of normal hemopoietic colony-forming cells. J. Cell. Physiol. 69, 177–184 (1967).
Wu, A. M., Till, J. E., Siminovitch, L. & McCulloch, E. A. Cytological evidence for a relationship between normal hemotopoietic colony-forming cells and cells of the lymphoid system. J. Exp. Med. 127, 455–464 (1968).
Abramson, S., Miller, R. G. & Phillips, R. A. The identification in adult bone marrow of pluripotent and restricted stem cells of the myeloid and lymphoid systems. J. Exp. Med. 145, 1567–1579 (1977).
Dick, J. E., Magli, M. C., Huszar, D., Phillips, R. A. & Bernstein, A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell 42, 71–79 (1985).
Keller, G., Paige, C., Gilboa, E. & Wagner, E. F. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature 318, 149–154 (1985).
Lemischka, I. R., Raulet, D. H. & Mulligan, R. C. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell 45, 917–927 (1986).
Walsh, C. & Cepko, C. L. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440 (1992).
Golden, J. A., Fields-Berry, S. C. & Cepko, C. L. Construction and characterization of a highly complex retroviral library for lineage analysis. Proc. Natl Acad. Sci. USA 92, 5704–5708 (1995).
Acknowledgements
The authors thank S. Naik and G. Bendle for valuable input during conception of this manuscript and members of the Schumacher laboratory for stimulating discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Parabiotic mice
-
Pairs of mice that are surgically joined by cutaneous vascular anastomosis so that they have a common blood circulation while maintaining separate organs and tissues.
- Intravital microscopy
-
Examination of biological processes by microscopy on living animals or viable explanted tissue.
- β-selection
-
The process leading to the proliferation and survival of thymocytes that have successfully recombined the β-chain of the T cell receptor locus to express a functional pre-T cell receptor on their cell surface.
- Cre–lox approach
-
A site-specific recombination system that is used to delete or rearrange genes by Cre recombinase activity. Two short DNA sequences (LoxP sites) are engineered to flank the target DNA. Depending on the orientation of the LoxP sites, expression of Cre recombinase leads to excision or inversion of the intervening sequence.
Rights and permissions
About this article
Cite this article
Schumacher, T., Gerlach, C. & van Heijst, J. Mapping the life histories of T cells. Nat Rev Immunol 10, 621–631 (2010). https://doi.org/10.1038/nri2822
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri2822
This article is cited by
-
Best Practices in Designing, Sequencing, and Identifying Random DNA Barcodes
Journal of Molecular Evolution (2023)
-
TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures
Nature Immunology (2019)
-
The regulation effect of AMPK in immune related diseases
Science China Life Sciences (2018)
-
Regulation of T cell immunity by cellular metabolism
Frontiers of Medicine (2018)
-
Reproducibility of Illumina platform deep sequencing errors allows accurate determination of DNA barcodes in cells
BMC Bioinformatics (2016)