Key Points
-
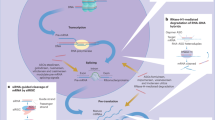
The key bottlenecks for small RNA drugs that harness RNA interference (RNAi) for selective gene knockdown in vivo include their delivery across the plasma membrane and their release from endosomes into the cytosol.
-
Small interfering RNA (siRNA)-based drugs can now be delivered into the cytosol of hepatocytes to suppress gene expression in the liver.
-
Recent siRNA clinical trials show durable and potent gene silencing in the liver, with manageable toxicity for a handful of disease targets.
-
The most effective strategies for gene knockdown in the liver use second-generation lipid nanoparticles or GalNAc-conjugated siRNAs that are taken up by the asialoglycoprotein receptor, which is exclusively expressed by hepatocytes.
-
Achieving gene knockdown outside the liver is still clinically unproven. The most attractive strategies use topical administration of siRNAs to accessible tissue sites, such as the skin, eye or mucosa, or use siRNAs that are covalently linked to an RNA aptamer that binds with high affinity to a cell surface receptor selectively expressed on cells being targeted for gene knockdown.
-
Methods that are being developed to deliver siRNAs therapeutically may eventually prove to be useful for delivering other nucleic acid therapeutics, including antisense oligonucleotides, mRNAs for gene expression, or CRISPR–Cas9 (clustered regularly interspaced short palindromic repeats–CRISPR-associated 9) for gene editing.
Abstract
Small interfering RNAs (siRNAs), which downregulate gene expression guided by sequence complementarity, can be used therapeutically to block the synthesis of disease-causing proteins. The main obstacle to siRNA drugs — their delivery into the target cell cytosol — has been overcome to allow suppression of liver gene expression. Here, we review the results of recent clinical trials of siRNA therapeutics, which show efficient and durable gene knockdown in the liver, with signs of promising clinical outcomes and little toxicity. We also discuss the barriers to more widespread applications that target tissues besides the liver and the most promising avenues to overcome them.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Zamecnik, P. C. & Stephenson, M. L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl Acad. Sci. USA 75, 280–284 (1978).
Stephenson, M. L. & Zamecnik, P. C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl Acad. Sci. USA 75, 285–288 (1978). First demonstration of the possibility of inhibiting gene expression using oligonucleotides.
Kole, R., Krainer, A. R. & Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11, 125–140 (2012).
Bennett, C. F. & Swayze, E. E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010).
Sharma, V. K., Sharma, R. K. & Singh, S. K. Antisense oligonucleotides: modifications and clinical trials. Med. Chem. Commun. 5, 1454–1471 (2014).
Raal, F. J. et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet 375, 998–1006 (2010).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). Initial report describing mediation of RNAi by double-stranded RNAs.
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001). Demonstration that exogenously supplied short double-stranded RNAs can specifically and efficiently inhibit gene expression in mammalian cells.
Song, E. et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9, 347–351 (2003). First proof of principle that siRNAs can be harnessed to treat disease in an in vivo model.
Morrissey, D. V. et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41, 1349–1356 (2005).
Chiu, Y.-L. & Rana, T. M. siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003).
Morrissey, D. V. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23, 1002–1007 (2005). First demonstration that siRNAs can be chemically modified to attenuate nuclease degradation and immune stimulation to allow for efficient lipid-based systemic administration.
Judge, A. D., Bola, G., Lee, A. C. H. & MacLachlan, I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13, 494–505 (2006).
Jackson, A. L. et al. Position-specific chemical modification of siRNAs reduces 'off-target' transcript silencing. RNA 12, 1197–1205 (2006).
Bartlett, D. W. & Davis, M. E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 34, 322–333 (2006).
Gilleron, J. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013).
Wittrup, A. et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. http://dx.doi.org/10.1038/nbt.3298 (2015).
Kanasty, R., Dorkin, J. R., Vegas, A. & Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977 (2013).
Zimmermann, T. S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006). First report of potent RNAi-mediated gene silencing in non-human primates after systemic administration of siRNA formulated in LNPs.
Akinc, A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 (2008).
Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28, 172–176 (2010).
Shi, B. et al. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. 59, 727–740 (2011).
Akinc, A. et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18, 1357–1364 (2010).
Judge, A. D. et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J. Clin. Invest. 119, 661–673 (2009).
Bartlett, D. W., Su, H., Hildebrandt, I. J., Weber, W. A. & Davis, M. E. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc. Natl Acad. Sci. USA 104, 15549–15554 (2007).
Song, E. et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 23, 709–717 (2005).
Peer, D., Zhu, P., Carman, C. V., Lieberman, J. & Shimaoka, M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc. Natl Acad. Sci. USA 104, 4095–4100 (2007).
Peer, D., Park, E. J., Morishita, Y., Carman, C. V. & Shimaoka, M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science 319, 627–630 (2008).
McNamara, J. O. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006).
Berezhnoy, A., Castro, I., Levay, A., Malek, T. R. & Gilboa, E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J. Clin. Invest. 124, 188–197 (2014).
Wheeler, L. A. et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest. 121, 2401–2412 (2011).
Davis, M. E. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010).
Nair, J. K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961 (2014). Description of triantennary GalNAc-conjugated siRNA for liver-targeted gene knockdown.
Wong, S. C. et al. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic Acid Ther. 22, 380–390 (2012).
Kortylewski, M. et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat. Biotechnol. 27, 925–932 (2009).
Soutschek, J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004).
Biessen, E. A. et al. Synthesis of cluster galactosides with high affinity for the hepatic asialoglycoprotein receptor. J. Med. Chem. 38, 1538–1546 (1995).
Matsuda, S. et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 10, 1181–1187 (2015).
Rajeev, K. G. et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 16, 903–908 (2015).
Manoharan, M. GalNAc-siRNA with enhanced stabilization chemistry: ESC-GalNAc-siRNA. Alnylam [online], (2014).
Thurston, T. L. M., Wandel, M. P., von Muhlinen, N., Foeglein, Á. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012).
Boussif, O., Zanta, M. A. & Behr, J. P. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Ther. 3, 1074–1080 (1996).
Rozema, D. B. et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl Acad. Sci. USA 104, 12982–12987 (2007).
Cohen, J. C., Boerwinkle, E., Mosley, T. H. Jr & Hobbs, H. H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Sorensen, B. et al. A subcutaneously administered RNAi therapeutic (ALN-AT3) targeting antithrombin for treatment of hemophilia: interim Phase 1 study results in healthy volunteers and patients with hemophilia A or B. Oral Poster Abstract 693. 56th American Society of Hematology [online], (2014).
Tabernero, J. et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 3, 406–417 (2013).
Reynolds, A. et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330 (2004).
Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199–213 (2002).
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Jackson, A. L. et al. Widespread siRNA 'off-target' transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 (2006).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Huang, L. et al. Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat. Biotechnol. 31, 350–356 (2013).
Judge, A. D. et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23, 457–462 (2005).
Kleinman, M. E. et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 452, 591–597 (2008).
Hornung, V. et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 (2005).
Poeck, H. et al. 5′-triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 14, 1256–1263 (2008).
Roberts, T. L., Sweet, M. J., Hume, D. A. & Stacey, K. J. Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J. Immunol. 174, 605–608 (2005).
Kedmi, R., Ben-Arie, N. & Peer, D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials 31, 6867–6875 (2010).
Landesman-Milo, D. & Peer, D. Toxicity profiling of several common RNAi-based nanomedicines: a comparative study. Drug Deliv. Transl. Res. 4, 96–103 (2014).
Maier, M. A. et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 21, 1570–1578 (2013).
Rusconi, C. Phase 3 evaluation of revolixys kit: study summary and lessons learned. Celebrating the 25th Anniversary of Selex, ASGCT meeting May 10–12, 2015, New Orleans (2015).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013). First demonstration of highly potent siRNA-mediated gene knockdown in humans.
Degenhardt, Y. & Lampkin, T. Targeting Polo-like kinase in cancer therapy. Clin. Cancer Res. 16, 384–389 (2010).
Geisbert, T. W. et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 375, 1896–1905 (2010).
Fitzgerald, K. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383, 60–68 (2014).
Adams, D. et al. Phase 2 open-label extension study of patisiran, an RNAi therapeutic for the treatment of familial amyloidotic polyneuropathy. Alnylam [online], (2014)
Zimmermann, T. et al. Phase I first-in-human trial of ALN-TTRsc, a novel RNA interference therapeutic for the treatment of familial amyloidotic cardiomyopathy (FAC). Alnylam [online], (2013).
Akinc, A. A. Subcutaneously administered investigational RNAi therapeutic (ALN-AT3) targeting antithrombin for treatment of hemophilia: interim Phase 1 study results in healthy volunteers and hemophilia A and B subjects. Alnylam [online], (2015).
[No authors listed.] RNAi Roundtable: ALN-PCSsc for the treatment of hypercholesterolemia. Alnylam [online], (2014).
Wooddell, C. I. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 21, 973–985 (2013).
Yuen, M.-F. et al. Phase II, dose ranging study of ARC-520, a siRNA-based therapeutic, in patients with chronic hepatitis B virus infection. American Association for the Study of Liver Diseases [online], (2014).
[No authors listed.] Arrowhead begins Phase 1 trial of ARC-AAT for treatment of liver disease associated with alpha-1 antitrypsin deficiency. Arrowhead Research [online], (2015).
Byrne, M. et al. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 29, 855–864 (2013).
[No authors listed.] Clinical trials — overview. RXI Pharmaceuticals [online]
Tolcher, A. W. et al. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J. Clin. Oncol. 33, 915_suppl 11006 (2015).
Dudek, H. et al. Knockdown of β-catenin with dicer-substrate siRNAs reduces liver tumor burden in vivo. Mol. Ther. 22, 92–101 (2014).
Schultheis, B. et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J. Clin. Oncol. 32, 4141–4148 (2014).
Wu, Y. et al. Durable protection from herpes simplex virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe 5, 84–94 (2009).
Nakayama, T. et al. Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Mol. Ther. 20, 1582–1589 (2012).
Alvarez-Erviti, L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345 (2011).
Kumar, P. et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 134, 577–586 (2008).
Yao, Y.-D. et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci. Transl Med. 4, 130ra48 (2012).
Cuellar, T. L. et al. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Res. 43, 1189–1203 (2015).
Rozema, D. B. et al. Protease-triggered siRNA delivery vehicles. J. Control. Release 209, 57–66 (2015).
Meade, B. R. et al. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 32, 1256–1261 (2014).
Warren, L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010).
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Goemans, N. M. et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N. Engl. J. Med. 364, 1513–1522 (2011).
Janssen, H. L. A. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013).
Bhat, B. et al. RG-101, a GalNAC-conjugated anti-miR employing aunique mechanism of action by targeting host factor microRNA-122 (miR-122), demonstrates potent activity and reduction of HCV in preclinical studies. Hepatology 58, 1393A (2013).
Hata, A. & Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 8, re3 (2015).
Daige, C. L. et al. Systemic delivery of a miR34a mimic as a potential therapeutic for liver cancer. Mol. Cancer Ther. 13, 2352–2360 (2014).
Prakash, T. P. et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 42, 8796–8807 (2014).
Rozema, D. B., Ekena, K., Lewis, D. L., Loomis, A. G. & Wolff, J. A. Endosomolysis by masking of a membrane-active agent (EMMA) for cytoplasmic release of macromolecules. Bioconjug. Chem. 14, 51–57 (2003).
Gilboa-Geffen, A. et al. Gene knockdown by EpCAM aptamer–siRNA chimeras suppresses epithelial breast cancers and their tumor-initiating cells. Mol. Cancer Ther. (in the press).
Acknowledgements
This work was supported by the Swedish Research Council (A.W.) and the US National Institutes of Health (NIH) grant CA139444 (to J.L.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.L. is on the Scientific Advisory Board of Alnylam Pharmaceuticals. A.W. declares no competing interests.
PowerPoint slides
Glossary
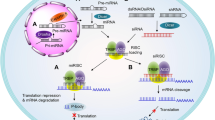
- RNA interference
-
(RNAi). An endogenous gene silencing mechanism, present in virtually all eukaryotic cells, by which short double-stranded RNA molecules induce translational inhibition and/or degradation of mRNAs containing partially complementary sequences.
- Gene knockdown
-
An experimental technique used to reduce gene expression using sequence-specific oligonucleotides, typically by RNA interference (RNAi) or antisense mechanisms.
- RNA-induced silencing complex
-
(RISC). The catalytic effector complex of RNA interference (RNAi)-mediated gene silencing. The RISC is a multiprotein complex that incorporates one strand of a small interfering RNA (siRNA) or microRNA.
- Aptamers
-
Oligonucleotides (DNA or RNA) selected to bind with high affinity to defined structures.
- MicroRNAs
-
(miRNAs). Endogenous, ~21-nucleotide-long, imperfectly paired double-stranded RNA molecules present in both plants and animals that guide the silencing of a multitude of genes bearing partially complementary sequences.
- Endosomal escape
-
The process of cytosolic entry of small interfering RNAs (siRNAs) from a vesicular compartment, following initial endocytosis of the siRNA (and delivery vehicle) into the target cell.
Rights and permissions
About this article
Cite this article
Wittrup, A., Lieberman, J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet 16, 543–552 (2015). https://doi.org/10.1038/nrg3978
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3978
This article is cited by
-
RNA interference in the era of nucleic acid therapeutics
Nature Biotechnology (2024)
-
Breaking the mold with RNA—a “RNAissance” of life science
npj Genomic Medicine (2024)
-
Biomembrane-derived nanoplexes for SiRNAs-pioneer innovation in delivery to lung adenocarcinoma
Journal of Nanoparticle Research (2024)
-
Engineering siRNA therapeutics: challenges and strategies
Journal of Nanobiotechnology (2023)
-
Amyotrophic lateral sclerosis: translating genetic discoveries into therapies
Nature Reviews Genetics (2023)