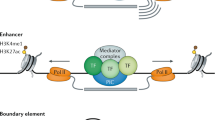
Key Points
-
Genetic studies during the past decades have revealed tightly regulated transcriptional networks that control robust developmental programs. However, this deterministic view of development contrasts with recent genomic studies that suggest that the multiple steps of transcription are by themselves rather leaky.
-
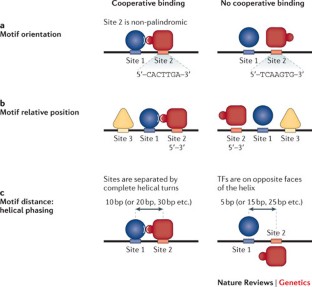
Although any single transcription factor (TF) can typically bind to thousands of sites throughout the genome, cis-regulatory activity at enhancers requires the concerted action of multiple binding events (which can be homotypic or heterotypic).
-
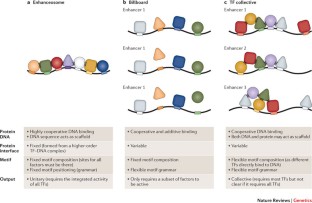
TFs bind to enhancers in a combinatorial manner, which is facilitated through direct and indirect cooperative mechanisms. The combinatorial nature of enhancer occupancy allows genes to be regulated in complex patterns in both space and time.
-
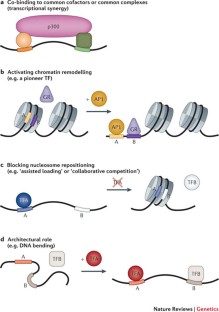
DNA and proteins can act together as a scaffold to cooperatively recruit TFs to enhancers.
-
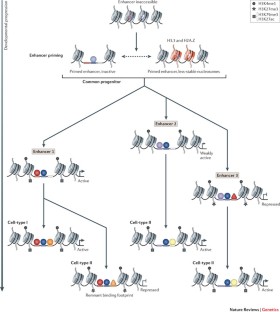
Enhancers undergo progressive changes during development, in which their occupancy by TFs, and the position of nucleosomes with or without post-translational modifications, reflect their inactive, poised or active state.
-
Pioneer TFs recruit chromatin-remodelling factors to reposition nucleosomes, thus facilitating the occupancy of other TFs at subsequent developmental stages.
-
Enhancer priming can occur through many different mechanisms. For example, some TF binding events may serve as placeholders to prevent nucleosome repositioning.
-
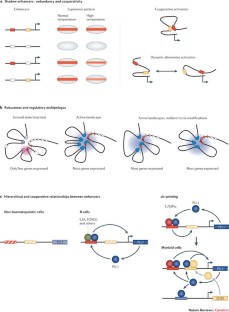
Partially redundant enhancers can act to buffer changes in environmental conditions to ensure robust developmental progression.
-
Multiple elements regulating the same gene seem to assemble in a three-dimensional structure with the promoter, and can have synergistic or repressive influences on each other.
Abstract
Developmental progression is driven by specific spatiotemporal domains of gene expression, which give rise to stereotypically patterned embryos even in the presence of environmental and genetic variation. Views of how transcription factors regulate gene expression are changing owing to recent genome-wide studies of transcription factor binding and RNA expression. Such studies reveal patterns that, at first glance, seem to contrast with the robustness of the developmental processes they encode. Here, we review our current knowledge of transcription factor function from genomic and genetic studies and discuss how different strategies, including extensive cooperative regulation (both direct and indirect), progressive priming of regulatory elements, and the integration of activities from multiple enhancers, confer specificity and robustness to transcriptional regulation during development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Davidson, E. H. Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010).
Peter, I. S. & Davidson, E. H. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (2011).
Lenhard, B., Sandelin, A. & Carninci, P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nature Rev. Genet. 13, 233–245 (2012).
Levine, M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010).
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Petrykowska, H. M., Vockley, C. M. & Elnitski, L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 18, 1238–1246 (2008).
Vokes, S. A., Ji, H., Wong, W. H. & McMahon, A. P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008).
Ayer, S. & Benyajati, C. Conserved enhancer and silencer elements responsible for differential Adh transcription in Drosophila cell lines. Mol. Cell. Biol. 10, 3512–3523 (1990).
Gaszner, M. & Felsenfeld, G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev. Genet. 7, 703–713 (2006).
Ohtsuki, S., Levine, M. & Cai, H. N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12, 547–556 (1998).
Calhoun, V. C., Stathopoulos, A. & Levine, M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl Acad. Sci. USA 99, 9243–9247 (2002).
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981).
Small, S., Blair, A. & Levine, M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047–4057 (1992).
Halfon, M. S. et al. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103, 63–74 (2000).
Yuh, C. H., Ransick, A., Martinez, P., Britten, R. J. & Davidson, E. H. Complexity and organization of DNA-protein interactions in the 5′-regulatory region of an endoderm-specific marker gene in the sea urchin embryo. Mech. Dev. 47, 165–186 (1994).
Sandmann, T. et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436–449 (2007).
Lettice, L. A. et al. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22, 459–467 (2012).
Stanojevic, D., Small, S. & Levine, M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254, 1385–1387 (1991).
Ip, Y. T., Levine, M. & Small, S. J. The bicoid and dorsal morphogens use a similar strategy to make stripes in the Drosophila embryo. J. Cell Sci. Suppl. 16, 33–38 (1992).
Xu, X., Yin, Z., Hudson, J. B., Ferguson, E. L. & Frasch, M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 12, 2354–2370 (1998).
Lee, H. H. & Frasch, M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development 132, 1429–1442 (2005).
Guss, K. A., Nelson, C. E., Hudson, A., Kraus, M. E. & Carroll, S. B. Control of a genetic regulatory network by a selector gene. Science 292, 1164–1167 (2001).
Mullen, A. C. et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565–576 (2011).
Trompouki, E. et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 (2011).
Sandmann, T. et al. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 10, 797–807 (2006).
Jakobsen, J. S. et al. Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev. 21, 2448–2460 (2007).
Cao, Y. et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18, 662–674 (2010).
Lin, Y. C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature Immunol. 11, 635–643 (2010).
Pilon, A. M. et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 118, e139–e148 (2011).
Gaudet, J. & Mango, S. E. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821–825 (2002).
Wilczynski, B. & Furlong, E. E. Dynamic CRM occupancy reflects a temporal map of developmental progression. Mol. Syst. Biol. 6, 383 (2010).
Yanez-Cuna, J. O., Dinh, H. Q., Kvon, E. Z., Shlyueva, D. & Stark, A. Uncovering cis-regulatory sequence requirements for context specific transcription factor binding. Genome Res. 25 Apr 2012 (doi:10.1101/gr.132811.111).
Zeitlinger, J. et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404 (2003).
Biggar, S. R. & Crabtree, G. R. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20, 3167–3176 (2001).
Giorgetti, L. et al. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol. Cell 37, 418–428 (2010).
Johnson, A. D., Meyer, B. J. & Ptashne, M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc. Natl Acad. Sci. USA 76, 5061–5065 (1979).
Lebrecht, D. et al. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proc. Natl Acad. Sci. USA 102, 13176–13181 (2005).
Zinzen, R. P., Senger, K., Levine, M. & Papatsenko, D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 16, 1358–1365 (2006).
Szymanski, P. & Levine, M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 14, 2229–2238 (1995).
Chopra, V. S. & Levine, M. Combinatorial patterning mechanisms in the Drosophila embryo. Brief. Funct. Genomic. Proteomic. 8, 243–249 (2009).
Masel, J. & Siegal, M. L. Robustness: mechanisms and consequences. Trends Genet. 25, 395–403 (2009).
Lin, Y. S., Carey, M., Ptashne, M. & Green, M. R. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature 345, 359–361 (1990).
Merika, M., Williams, A. J., Chen, G., Collins, T. & Thanos, D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287 (1998).
Voss, T. C. et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554 (2011).
Miller, J. A. & Widom, J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 23, 1623–1632 (2003).
Panne, D., Maniatis, T. & Harrison, S. C. An atomic model of the interferon-β enhanceosome. Cell 129, 1111–1123 (2007).
Falvo, J. V., Thanos, D. & Maniatis, T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell 83, 1101–1111 (1995).
Siggers, T., Duyzend, M. H., Reddy, J., Khan, S. & Bulyk, M. L. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol. Syst. Biol. 7, 555 (2011). This is an interesting study demonstrating that the binding of a cofactor that does not contain a DNA-binding domain can alter the sequence specificity of a transcription factor complex.
Noyes, M. B. et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277–1289 (2008).
Carr, A. & Biggin, M. D. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J. 18, 1598–1608 (1999).
Slattery, M. et al. Cofactor binding evokes latent differences in DNA binding specificity between hox proteins. Cell 147, 1270–1282 (2011).
Liu, X., Lee, C. K., Granek, J. A., Clarke, N. D. & Lieb, J. D. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 16, 1517–1528 (2006).
Li, X.-y. et al. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 12, R34 (2011). The authors found that TF occupancy correlates better with DNA accessibility (estimated by DNaseI hypersensitivity) than with predicted sequence affinity, suggesting that this simple property could explain the clustering of TF binding.
John, S. et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genet. 43, 264–268 (2011).
Degner, J. F. et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 (2012).
Buck, M. J. & Lieb, J. D. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nature Genet. 38, 1446–1451 (2006).
Carr, A. & Biggin, M. D. Accessibility of transcriptionally inactive genes is specifically reduced at homeoprotein-DNA binding sites in Drosophila. Nucleic Acids Res. 28, 2839–2846 (2000).
John, S. et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29, 611–624 (2008).
Vicent, G. P. et al. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 5, e1000567 (2009).
Hartley, P. D. & Madhani, H. D. Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458 (2009).
Yarragudi, A., Miyake, T., Li, R. & Morse, R. H. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 9152–9164 (2004).
Workman, J. L. & Kingston, R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science 258, 1780–1784 (1992).
Morse, R. H. Nucleosome disruption by transcription factor binding in yeast. Science 262, 1563–1566 (1993).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 41, 376–381 (2009).
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Heintzman, N. D. & Ren, B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541–549 (2009).
Ong, C. T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Rev. Genet. 12, 283–293 (2011).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011). References 71 and 72 identified a strong correlation between the presence of H3K27ac on cis -regulatory elements and the activity of the neighbouring gene, providing the first indication that this mark, rather than H3K4me1, is a good indicator of active regulatory elements.
Bonn, S. et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genet. 44, 148–156 (2012).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
McManus, S. et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 30, 2388–2404 (2011).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Mercer, E. M. et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity 35, 413–425 (2011).
Flames, N. & Hobert, O. Gene regulatory logic of dopamine neuron differentiation. Nature 458, 885–889 (2009).
Zinzen, R. P., Girardot, C., Gagneur, J., Braun, M. & Furlong, E. E. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009).
Brody, T. et al. Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers. Dev. Dyn. 241, 169–189 (2012).
Kaplan, T. et al. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 7, e1001290 (2011).
Narlikar, L. et al. Genome-wide discovery of human heart enhancers. Genome Res. 20, 381–392 (2010).
Senger, K. et al. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13, 19–32 (2004).
Swanson, C. I., Evans, N. C. & Barolo, S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370 (2010). By carrying out an extensive mutagenesis analysis, this elegant study found that the sequence of a D. melanogaster enhancer is highly constrained, with the presence and relative positioning of almost all sequence motifs being essential for some aspect of the enhancer's activity. Interestingly, in other Drosophila species, the homologous enhancer adopted different 'solutions' to give the same activity.
Thanos, D. & Maniatis, T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 (1995).
Merika, M. & Thanos, D. Enhanceosomes. Curr. Opin. Genet. Dev. 11, 205–208 (2001).
Kulkarni, M. M. & Arnosti, D. N. Information display by transcriptional enhancers. Development 130, 6569–6575 (2003).
Arnosti, D. N. & Kulkarni, M. M. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890–898 (2005).
Brown, C. D., Johnson, D. S. & Sidow, A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317, 1557–1560 (2007).
Liberman, L. M. & Stathopoulos, A. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol. 327, 578–589 (2009).
Junion, G. et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (2012).
Gao, F., Foat, B. C. & Bussemaker, H. J. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics 5, 31 (2004).
Ucar, D., Beyer, A., Parthasarathy, S. & Workman, C. T. Predicting functionality of protein-DNA interactions by integrating diverse evidence. Bioinformatics 25, i137–i144 (2009).
Krejci, A., Bernard, F., Housden, B. E., Collins, S. & Bray, S. J. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2, ra1 (2009).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Biswas, A. K. & Johnson, D. G. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 72, 13–17 (2012).
Lickwar, C. R., Mueller, F., Hanlon, S. E., McNally, J. G. & Lieb, J. D. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484, 251–255 (2012). The authors tagged a transcription factor with two different tags and carried out a competitive ChIP assay to measure how long it took for the newly transcribed tagged TF to outcompete its alternatively tagged counterpart for chromatin binding. The resulting information provides a nice way to measure TF residence time on each site throughout the genome, which seems to be a better indicator of functional binding events than static occupancy data.
Tagoh, H. et al. Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J. 23, 4275–4285 (2004).
Krysinska, H. et al. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol. Cell. Biol. 27, 878–887 (2007).
Almer, A., Rudolph, H., Hinnen, A. & Horz, W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 (1986).
Biddie, S. C. et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 (2011).
de la Serna, I. L. et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997–4009 (2005).
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008). This study shows that FOXA1 occupancy correlates with the prior presence of H3K4me2 at specific enhancer elements. This histone modification directs FOXA1 to induce further chromatin remodelling and TF occupancy specifically at those enhancers. This suggests that even what is perceived as a pioneer factor requires a prior chromatin context.
Siersbæk, R. et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 30, 1459–1472 (2011).
Liber, D. et al. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell 7, 114–126 (2010).
Xu, J. et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838 (2009).
Hoogenkamp, M. et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114, 299–309 (2009).
Liang, H. L. et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008).
Nien, C. Y. et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339 (2011).
Harrison, M. M., Li, X.-y., Kaplan, T., Botchan, M. R. & Eisen, M. B. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266 (2011). References 109 and 110 indicate that Zelda is a major global regulator of the MZT in D. melanogaster . Zelda binds to thousands of regulatory elements prior to the MZT and prior to the occupancy of other known factors and is required for the zygotic gene activation of a large number of genes.
Struffi, P. et al. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3 + 7 enhancer. Development 138, 4291–4299 (2011).
Bell, O., Tiwari, V. K., Thoma, N. H. & Schubeler, D. Determinants and dynamics of genome accessibility. Nature Rev. Genet. 12, 554–564 (2011).
Xu, J. et al. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 12377–12382 (2007).
Serandour, A. A. et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565 (2011).
Stadler, M. B. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 (2011).
Pevny, L. & Placzek, M. SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 15, 7–13 (2005).
Olson, E. N. & Klein, W. H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8, 1–8 (1994).
Bergsland, M. et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453–2464 (2011).
Cui, K. et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4, 80–93 (2009).
Watts, J. A. et al. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 7, e1002277 (2011).
Zeitlinger, J. et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385–390 (2007).
Hong, J. W., Hendrix, D. A. & Levine, M. S. Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 (2008).
Barolo, S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–141 (2012).
Frankel, N. et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010).
Perry, M. W., Boettiger, A. N., Bothma, J. P. & Levine, M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562–1567 (2010). Developmental genes often have multiple enhancers that seem to have overlapping activities and functions under normal developmental conditions. References 124 and 125 showed that the removal of one (or both) of the enhancers leads to developmental defects when the environmental conditions are more extreme, indicating that 'shadow' enhancers provide robustness to gene expression in the midst of fluctuating environmental temperatures.
Barolo, S. & Levine, M. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16, 2883–2891 (1997).
Perry, M. W., Boettiger, A. N. & Levine, M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl Acad. Sci. USA 108, 13570–13575 (2011).
Yao, L.-C. et al. Multiple modular promoter elements drive graded brinker expression in response to the Dpp morphogen gradient. Development 135, 2183–2192 (2008).
Prazak, L., Fujioka, M. & Gergen, J. P. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev. Biol. 344, 1048–1059 (2010).
Spitz, F., Gonzalez, F. & Duboule, D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405–417 (2003).
Gonzalez, F., Duboule, D. & Spitz, F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 306, 847–859 (2007).
Montavon, T. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). Genetic and transgenic analyses showed that mouse HoxD genes are controlled by many remote regulatory elements that are separated by large distances. Chromatin conformation capture experiments revealed that these elements come together in cells where they contribute to HoxD gene expression, providing a three-dimensional view of the complex folding of regulatory landscapes during embryonic development.
Suter, D. M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011).
Amano, T. et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 (2008).
Jeong, Y., El-Jaick, K., Roessler, E., Muenke, M. & Epstein, D. J. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772 (2006).
Leddin, M. et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 117, 2827–2838 (2011).
Jack, J. & DeLotto, Y. Structure and regulation of a complex locus: the cut gene of Drosophila. Genetics 139, 1689–1700 (1995).
Wunderle, V. M., Critcher, R., Hastie, N., Goodfellow, P. N. & Schedl, A. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc. Natl Acad. Sci. USA 95, 10649–10654 (1998).
Sagai, T., Hosoya, M., Mizushina, Y., Tamura, M. & Shiroishi, T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132, 797–803 (2005).
Visel, A., Rubin, E. M. & Pennacchio, L. A. Genomic views of distant-acting enhancers. Nature 461, 199–205 (2009).
Splinter, E. & de Laat, W. The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J. 30, 4345–4355 (2011).
Butler, J. E. & Kadonaga, J. T. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519 (2001).
Zuniga, A. et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 18, 1553–1564 (2004).
Cajiao, I., Zhang, A., Yoo, E. J., Cooke, N. E. & Liebhaber, S. A. Bystander gene activation by a locus control region. EMBO J. 23, 3854–3863 (2004).
Lower, K. M. et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc. Natl Acad. Sci. USA 106, 21771–21776 (2009).
Ruf, S. et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nature Genet. 43, 379–386 (2011).
Klopocki, E. et al. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 45, 370–375 (2008).
Kurth, I. et al. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nature Genet. 41, 862–863 (2009).
Dathe, K. et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 84, 483–492 (2009).
Cande, J., Goltsev, Y. & Levine, M. S. Conservation of enhancer location in divergent insects. Proc. Natl Acad. Sci. USA 106, 14414–14419 (2009).
Mongin, E., Dewar, K. & Blanchette, M. Long-range regulation is a major driving force in maintaining genome integrity. BMC Evol. Biol. 9, 203 (2009).
Engstrom, P. G., Ho Sui, S. J., Drivenes, O., Becker, T. S. & Lenhard, B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 17, 1898–1908 (2007).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Ludwig, M. Z., Manu, Kittler, R., White, K. P. & Kreitman, M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 7, e1002364 (2011).
Janssens, H. et al. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nature Genet. 38, 1159–1165 (2006).
Klingler, M., Soong, J., Butler, B. & Gergen, J. P. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev. Biol. 177, 73–84 (1996).
MacArthur, S. et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80 (2009).
Liu, Y. H. et al. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev. Cell 16, 280–291 (2009).
Zheng, J. et al. Epistatic relationships reveal the functional organization of yeast transcription factors. Mol. Syst. Biol. 6, 420 (2010).
Acknowledgements
We are very grateful to J. Erceg, D. Harnett and P. Khoueiry for useful comments, with particular thanks to R. Zinzen and D. Garfield for some rephrasing. We apologize to all colleagues whose work was omitted due to space limitations; we unfortunately could not comment on all the interesting papers in the field. F.S. is supported by grants from the Human Frontier Science Program (HFSP), a European Commission grant (EC-FP7, grant Health 223210/CISSTEM). E.E.M.F. is supported by grants from the HFSP, the European Research Area for Systems Biology (ERASysBio) for the ModHeart project and the German Research Foundation (DFG; grant FU 750/1-1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Core promoter
-
The region of a gene to which RNA polymerase II and the general transcription factors (GTFs) bind to initiate transcription. Core promoters span ~40 base pairs upstream and downstream of the transcription start site.
- Silencers
-
DNA sequences to which repressor factors bind and mediate the silencing of promoters through interactions with the basal transcriptional machinery or enhancers.
- Insulators
-
Chromatin elements that act as barriers against the influence of positive signals (from enhancers) or negative signals (from silencers and from heterochromatin).
- Tethering elements
-
Cis-regulatory elements that contribute to directing a remote enhancer activity towards a specific gene among the surrounding genes. They are usually close to the promoter-proximal region of the target gene.
- Bone morphogenetic protein
-
(BMP). A group of molecules of the transforming growth factor-α (TGFα) family that can induce bone formation and ventralize the vertebrate embryo. Decapentaplegic (DPP) is the fly homologue of the vertebrate BMPs.
- Bicoid
-
A maternally deposited Drosophila melanogaster transcription factor that is localized at the anterior end of the embryo and forms a concentration gradient that decreases from the anterior to posterior. This transcription factor regulates transcription at different concentrations along its expression gradient, which is essential for establishing the segmentation of the early blastoderm embryo.
- Mediator
-
The ~30-subunit co-activator complex that is necessary for successful transcription at class II promoters of metazoan genes. Mediator coordinates the signals between enhancers and the general transcription machinery through its interaction with RNA polymerase II and site-specific factors.
- SAGA
-
A multiprotein complex involved in the regulation of transcription that possesses histone acetyltransferase and TATA-box-binding protein (TBP)-binding activities. The budding-yeast complex includes Gcn5, several proteins of the Spt and Ada families, and TBP-associated factors (TAFs); analogous complexes in other species have analogous compositions.
- Nucleosome-depleted regions
-
(NDRs). DNA is usually wrapped around histone octamers, forming nucleosomes that contribute to genome compaction in the nucleus. The distribution of nucleosomes is variable and regulated, and some genomic regions, termed NDRs, show a lower density or absence of nucleosomes.
- Chromatin immunoprecipitation
-
(ChIP). A technique for identifying potential regulatory sequences that are bound by a protein of interest. Covalently crosslinked DNA–chromatin extracts (complexes of DNA and protein) are isolated using antibodies that recognize specific DNA-binding proteins. In ChIP–chip, the ChIP step is followed by microarray analysis, whereas in ChIP–seq, it is followed by high-throughput sequencing.
- Maternal-to-zygotic transition
-
(MZT). A period of embryonic development that coincides with the transcriptional activation of the zygotic (embryonic) genome. Most of the RNA and proteins that are present in the embryo before the MZT are provided by the mother, through maternal loading into the oocyte.
- TAGteam motifs
-
A collection of related heptamer motifs (consensus sequence CAGGTAG) that are enriched in the vicinity of genes that are transcribed during the early blastoderm stages of Drosophila melanogaster embryogenesis; these motifs are recognized by the transcription factor Zelda.
- Splanchnic mesoderm
-
The part of the mesoderm that associates with the endoderm and that will develop later into gut muscles and heart. In Drosophila melanogaster the splanchnic mesoderm is located dorsally, whereas in vertebrates it is ventral.
Rights and permissions
About this article
Cite this article
Spitz, F., Furlong, E. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13, 613–626 (2012). https://doi.org/10.1038/nrg3207
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3207
This article is cited by
-
Genome-scale identification and comparative analysis of transcription factors in thermophilic cyanobacteria
BMC Genomics (2024)
-
Affinity-optimizing enhancer variants disrupt development
Nature (2024)
-
Transcription factor ATMIN facilitates chemoresistance in nasopharyngeal carcinoma
Cell Death & Disease (2024)
-
3D Enhancer–promoter networks provide predictive features for gene expression and coregulation in early embryonic lineages
Nature Structural & Molecular Biology (2024)
-
A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development
Nature Communications (2024)