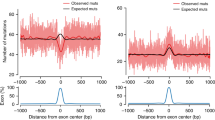
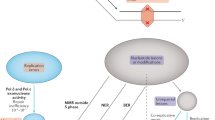
Key Points
-
Recent technological advances have made it possible to directly measure the frequency of rare nucleotide substitution mutations in human germline DNA. Semen is an ideal source of genetic material as a single sample can contain over 108 sperm.
-
Some human nucleotides have mutation frequencies that are orders of magnitude greater than the genome average and, at least in some disease-causing examples, there is evidence that this increased frequency is not due to more frequent mutation. Rather, a selective advantage conferred on the male germline cells by the mutation has been suggested; other diseases may also achieve high frequencies in the population owing to germline selection.
-
The indirect method of studying nucleotide substitution mutations by comparing aligned sequences in different species has found that the mutation rate varies across the genome. Many genomic factors such as coding versus non-coding sequence, base identity, GC content, recombination rate, and proximity to insertions or deletions are correlated with this rate, although it is likely that no single factor can explain all the variation.
-
Both interspecific sequence comparisons and analysis of parental origins of human disease mutations in families suggest that nucleotide substitutions occur more frequently in males than females. Recent studies have shown that mutations at CpG sites are significantly more male-biased if the CpG sites are in CpG islands.
-
Human disease mutations increase in frequency with the father's age; this has long been thought to result from the life-long divisions of the male germ cells. In some cases, new evidence suggests that selection of germ cells carrying the new mutation can explain the age-dependent increase.
Abstract
Surprising findings about human germline mutation have come from applying new technologies to detect rare mutations in germline DNA, from analysing DNA sequence divergence between humans and closely related species, and from investigating human polymorphic variation. In this Review we discuss how these approaches affect our current understanding of the roles of sex, age, mutation hot spots, germline selection and genomic factors in determining human nucleotide substitution mutation patterns and frequencies. To enhance our understanding of mutation and disease, more extensive molecular data on the human germ line with regard to mutation origin, DNA repair, epigenetic status and the effect of newly arisen mutations on gamete development are needed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Weatherall, D. J. The global problem of genetic disease. Ann. Hum. Biol. 32, 117–122 (2005).
Hassold, T., Hall, H. & Hunt, P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16, R203–R208 (2007).
Pacchierotti, F., Adler, I. D., Eichenlaub-Ritter, U. & Mailhes, J. B. Gender effects on the incidence of aneuploidy in mammalian germ cells. Environ. Res. 104, 46–69 (2007).
Rosenbusch, B. E. Mechanisms giving rise to triploid zygotes during assisted reproduction. Fertil. Steril. 90, 49–55 (2008).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nature Rev. Genet. 5, 435–445 (2004).
Kelkar, Y. D., Tyekucheva, S., Chiaromonte, F. & Makova, K. D. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 18, 30–38 (2008).
Bois, P. & Jeffreys, A. J. Minisatellite instability and germline mutation. Cell. Mol. Life Sci. 55, 1636–1648 (1999).
Lupski, J. R. & Stankiewicz, P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 1, e49 (2005).
Inoue, K. & Lupski, J. R. Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet. 3, 199–242 (2002).
Sharp, A. J., Cheng, Z. & Eichler, E. E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 7, 407–442 (2006).
Sen, S. K. et al. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 79, 41–53 (2006).
Scherer, S. W. et al. Challenges and standards in integrating surveys of structural variation. Nature Genet. 39 (Suppl.), S7–S15 (2007).
Turner, D. J. et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nature Genet. 40, 90–95 (2008).
Babushok, D. V. & Kazazian, H. H. Jr. Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28, 527–539 (2007).
Chen, J. M., Stenson, P. D., Cooper, D. N. & Ferec, C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum. Genet. 117, 411–427 (2005).
Cordaux, R., Hedges, D. J., Herke, S. W. & Batzer, M. A. Estimating the retrotransposition rate of human Alu elements. Gene 373, 134–137 (2006).
Jacobs, P. A., Browne, C., Gregson, N., Joyce, C. & White, H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J. Med. Genet. 29, 103–108 (1992).
Baptista, J. et al. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am. J. Hum. Genet. 82, 927–936 (2008).
Chen, J.-M., Cooper, D. N., Chuzhanova, N., Ferec, C. & Patrinos, G. P. Gene conversion: mechanisms, evolution and human disease. Nature Rev. Genet. 8, 762–775 (2007).
Vogel, F. & Motulsky, A. G. Human Genetics: Problems and Approaches (Springer, Berlin, 1997).
Crow, J. F. The origins, patterns and implications of human spontaneous mutation. Nature Rev. Genet. 1, 40–47 (2000).
Glaser, R. L. et al. The paternal-age effect in Apert syndrome is due, in part, to the increased frequency of mutations in sperm. Am. J. Hum. Genet. 73, 939–947 (2003).
Tiemann-Boege, I. et al. The observed human sperm mutation frequency cannot explain the achondroplasia paternal age effect. Proc. Natl Acad. Sci. USA 99, 14952–14957 (2002).
Goriely, A., McVean, G. A., Rojmyr, M., Ingemarsson, B. & Wilkie, A. O. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science 301, 643–646 (2003).
Cole, D. N., Carlson, J. A. & Wilson, V. L. Human germline and somatic cells have similar TP53 and Kirsten-RAS gene single base mutation frequencies. Environ. Mol. Mutagen. 49, 417–425 (2008).
Choi, S. K., Yoon, S. R., Calabrese, P. & Arnheim, N. A germ-line-selective advantage rather than an increased mutation rate can explain some unexpectedly common human disease mutations. Proc. Natl Acad. Sci. USA 105, 10143–10148 (2008).
Qin, J. et al. The molecular anatomy of spontaneous germline mutations in human testes. PLoS Biol. 5, e224 (2007).
Liu, Q. & Sommer, S. S. Detection of extremely rare alleles by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification (Bi-PAP-A): measurement of mutation load in mammalian tissues. Biotechniques 36, 156–166 (2004).
Nachman, M. W. & Crowell, S. L. Estimate of the mutation rate per nucleotide in humans. Genetics 156, 297–304 (2000).
The Chimpanzee Sequencing and Analysis Consortium.Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005).
Ebersberger, I., Metzler, D., Schwarz, C. & Paabo, S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70, 1490–1497 (2002).
Chen, F. C. & Li, W. H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68, 444–456 (2001).
Kondrashov, A. S. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum. Mutat. 21, 12–27 (2003).
Orioli, I. M., Castilla, E. E., Scarano, G. & Mastroiacovo, P. Effect of paternal age in achondroplasia, thanatophoric dysplasia, and osteogenesis imperfecta. Am. J. Med. Genet. 59, 209–217 (1995).
Horton, W. A., Hall, J. G. & Hecht, J. T. Achondroplasia. Lancet 370, 162–172 (2007).
Cohen, M. M. et al. Birth prevalence study of the Apert syndrome. Am. J. Med. Genet. 42, 655–659 (1992).
Tolarova, M. M., Harris, J. A., Ordway, D. E. & Vargervik, K. Birth prevalence, mutation rate, sex ratio, parents' age, and ethnicity in Apert syndrome. Am. J. Med. Genet. 72, 394–398 (1997).
Rousseau, F. et al. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature 371, 252–254 (1994).
Shiang, R. et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell 78, 335–342 (1994).
Bellus, G. A. et al. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 56, 368–373 (1995).
Park, W. J. et al. Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. Am. J. Hum. Genet. 57, 321–328 (1995).
Wilkie, A. O. et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nature Genet. 9, 165–172 (1995).
Goriely, A. et al. Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc. Natl Acad. Sci. USA 102, 6051–6056 (2005).
Crow, J. F. Age and sex effects on human mutation rates: an old problem with new complexities. J. Radiat. Res. 47 (Suppl. B), B75–B82 (2006).
Kan, S. H. et al. Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am. J. Hum. Genet. 70, 472–486 (2002).
Morrison, S. J. & Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074 (2006).
Knoblich, J. A. Mechanisms of asymmetric stem cell division. Cell 132, 583–597 (2008).
Dakouane Giudicelli, M. et al. Increased achondroplasia mutation frequency with advanced age and evidence for G1138A mosaicism in human testis biopsies. Fertil. Steril. 89, 1651–1656 (2007).
Eswarakumar, V. P., Lax, I. & Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 1139–1149 (2005).
Thisse, B. & Thisse, C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 287, 390–402 (2005).
Runeberg-Roos, P. & Saarma, M. Neurotrophic factor receptor RET: structure, cell biology, and inherited diseases. Ann. Med. 39, 572–580 (2007).
Carlson, K. M. et al. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc. Natl Acad. Sci. USA 91, 1579–1583 (1994).
Carlson, K. M. et al. Parent-of-origin effects in multiple endocrine neoplasia type 2B. Am. J. Hum. Genet. 55, 1076–1082 (1994).
Eng, C. et al. Point mutation within the tyrosine kinase domain of the RET proto-oncogene in multiple endocrine neoplasia type 2B and related sporadic tumours. Hum. Mol. Genet. 3, 237–241 (1994).
Wray, C. J. et al. Failure to recognize multiple endocrine neoplasia 2B: more common than we think? Ann. Surg. Oncol. 15, 293–301 (2008).
Oatley, J. M. & Brinster, R. L. Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263–286 (2008).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 23, 185–188 (1999).
Bird, A. The methyl-CpG-binding protein MeCP2 and neurological disease. Biochem. Soc. Trans. 36, 575–583 (2008).
Percy, A. K. et al. Rett syndrome: North American database. J. Child. Neurol. 22, 1338–1341 (2007).
Trappe, R. et al. MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am. J. Hum. Genet. 68, 1093–1101 (2001).
Malter, H. E. et al. Characterization of the full fragile-X-syndrome mutation in fetal gametes. Nature Genet. 15, 165–169 (1997).
Moutou, C., Vincent, M. C., Biancalana, V. & Mandel, J. L. Transition from premutation to full mutation in fragile X syndrome is likely to be prezygotic. Hum. Mol. Genet. 6, 971–979 (1997).
Temmerman, N. D. et al. Intergenerational instability of the expanded CTG repeat in the DMPK gene: studies in human gametes and preimplantation embryos. Am. J. Hum. Genet. 75, 325–329 (2004).
Moseley, M. L. et al. SCA8 CTG repeat: en masse contractions in sperm and intergenerational sequence changes may play a role in reduced penetrance. Hum. Mol. Genet. 9, 2125–2130 (2000).
Silveira, I. et al. High germinal instability of the (CTG)n at the SCA8 locus of both expanded and normal alleles. Am. J. Hum. Genet. 66, 830–840 (2000).
De Michele, G. et al. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 7, 1901–1906 (1998).
Delatycki, M. B. et al. Sperm DNA analysis in a Friedreich ataxia premutation carrier suggests both meiotic and mitotic expansion in the FRDA gene. J. Med. Genet. 35, 713–716 (1998).
Salat, U., Bardoni, B., Wohrle, D. & Steinbach, P. Increase of FMRP expression, raised levels of FMR1 mRNA, and clonal selection in proliferating cells with unmethylated fragile X repeat expansions: a clue to the sex bias in the transmission of full mutations? J. Med. Genet. 37, 842–850 (2000).
Hulten, M. A. et al. On the origin of trisomy 21 Down syndrome. Mol. Cytogenet. 1, 21 (2008).
Hastings, I. M. Germline selection: population genetic aspects of the sexual/asexual life cycle. Genetics 129, 1167–1176 (1991).
Smith, N. G., Webster, M. T. & Ellegren, H. Deterministic mutation rate variation in the human genome. Genome Res. 12, 1350–1356 (2002).
Hellmann, I. et al. Why do human diversity levels vary at a megabase scale? Genome Res. 15, 1222–1231 (2005).
Ellegren, H., Smith, N. G. & Webster, M. T. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 13, 562–568 (2003).
Green, P., Ewing, B., Miller, W., Thomas, P. J. & Green, E. D. Transcription-associated mutational asymmetry in mammalian evolution. Nature Genet. 33, 514–517 (2003).
Touchon, M., Arneodo, A., d'Aubenton-Carafa, Y. & Thermes, C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 32, 4969–4978 (2004).
Hanawalt, P. C. & Spivak, G. Transcription-coupled DNA repair: two decades of progress and surprises. 9, 958–970 (2008).
Polak, P. & Arndt, P. F. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 18, 1216–1223 (2008).
Majewski, J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 73, 688–692 (2003).
Webster, M. T., Smith, N. G., Lercher, M. J. & Ellegren, H. Gene expression, synteny, and local similarity in human noncoding mutation rates. Mol. Biol. Evol. 21, 1820–1830 (2004).
Anagnostopoulos, T., Green, P. M., Rowley, G., Lewis, C. M. & Giannelli, F. DNA variation in a 5-Mb region of the X chromosome and estimates of sex-specific/type-specific mutation rates. Am. J. Hum. Genet. 64, 508–517 (1999).
Krawczak, M., Ball, E. V. & Cooper, D. N. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 63, 474–488 (1998).
Cooper, D. N. & Krawczak, M. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum. Genet. 85, 55–74 (1990).
Hwang, D. G. & Green, P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl Acad. Sci. USA 101, 13994–14001 (2004).
Arndt, P. F. & Hwa, T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics 21, 2322–2328 (2005).
Hess, S. T., Blake, J. D. & Blake, R. D. Wide variations in neighbor-dependent substitution rates. J. Mol. Biol. 236, 1022–1033 (1994).
Zhao, Z. & Boerwinkle, E. Neighboring-nucleotide effects on single nucleotide polymorphisms: a study of 2.6 million polymorphisms across the human genome. Genome Res. 12, 1679–1686 (2002).
Hodgkinson, A., Ladoukakis, E. & Eyre-Walker, A. Cryptic variation in the human mutation rate. PLoS Biol. 7, e1000027 (2009).
Jeffreys, A. J. & Neumann, R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nature Genet. 31, 267–271 (2002).
Duret, L. & Arndt, P. F. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4, e1000071 (2008).
Hardison, R. C. et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13, 13–26 (2003).
Walser, J. C., Ponger, L. & Furano, A. V. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res. 18, 1403–1414 (2008).
Lercher, M. J. & Hurst, L. D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18, 337–340 (2002).
Myers, S. et al. The distribution and causes of meiotic recombination in the human genome. Biochem. Soc. Trans. 34, 526–530 (2006).
Spencer, C. C. et al. The influence of recombination on human genetic diversity. PLoS Genet. 2, e148 (2006).
Arnheim, N., Calabrese, P. & Tiemann-Boege, I. Mammalian meiotic recombination hot spots. Annu. Rev. Genet. 41, 369–399 (2007).
Tyekucheva, S. et al. Human–macaque comparisons illuminate variation in neutral substitution rates. Genome Biol. 9, R76 (2008).
Tian, D. et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455, 105–108 (2008).
Honma, M. et al. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair 6, 781–788 (2007).
Rattray, A. J., Shafer, B. K., McGill, C. B. & Strathern, J. N. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics 162, 1063–1077 (2002).
Prendergast, J. G. et al. Chromatin structure and evolution in the human genome. BMC Evol. Biol. 7, 72 (2007).
Haldane, J. B. S. The mutation rate of the gene for haemophilia and its segregation ratios in males and females. Ann. Eugen. 13, 262–271 (1947).
Hurst, L. D. & Ellegren, H. Sex biases in the mutation rate. Trends Genet. 14, 446–452 (1998).
Glaser, R. L. & Jabs, E. W. Dear old dad. Sci. Aging Knowledge Environ. 2004, re1 (2004).
Li, W. H., Yi, S. & Makova, K. Male-driven evolution. Curr. Opin. Genet. Dev. 12, 650–656 (2002).
Ellegren, H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc. Biol. Sci. 274, 1–10 (2007).
Grimm, T. et al. On the origin of deletions and point mutations in Duchenne muscular dystrophy: most deletions arise in oogenesis and most point mutations result from events in spermatogenesis. J. Med. Genet. 31, 183–186 (1994).
Becker, J. et al. Characterization of the factor VIII defect in 147 patients with sporadic hemophilia A: family studies indicate a mutation type-dependent sex ratio of mutation frequencies. Am. J. Hum. Genet. 58, 657–670 (1996).
Rossiter, J. P. et al. Factor VIII gene inversions causing severe hemophilia A originate almost exclusively in male germ cells. Hum. Mol. Genet. 3, 1035–1039 (1994).
Kluwe, L. et al. The parental origin of new mutations in neurofibromatosis 2. Neurogenetics 3, 17–24 (2000).
Sommer, S. S., Scaringe, W. A. & Hill, K. A. Human germline mutation in the factor IX gene. Mutat. Res. 487, 1–17 (2001).
Zlotogora, J. Germ line mosaicism. Hum. Genet. 102, 381–386 (1998).
Youssoufian, H. & Pyeritz, R. E. Mechanisms and consequences of somatic mosaicism in humans. Nature Rev. Genet. 3, 748–758 (2002).
Erickson, R. P. Somatic gene mutation and human disease other than cancer. Mutat. Res. 543, 125–136 (2003).
Hall, J. G. Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am. J. Hum. Genet. 43, 355–363 (1988).
Sippel, K. C. et al. Frequency of somatic and germ-line mosaicism in retinoblastoma: implications for genetic counseling. Am. J. Hum. Genet. 62, 610–619 (1998).
Leuer, M. et al. Somatic mosaicism in hemophilia A: a fairly common event. Am. J. Hum. Genet. 69, 75–87 (2001).
Evans, D. G. et al. Mosaicism in neurofibromatosis type 2: an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including multiple ligation-dependent probe amplification. J. Med. Genet. 44, 424–428 (2007).
Kehrer-Sawatzki, H. & Cooper, D. N. Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J. Med. Genet. 45, 622–631 (2008).
Winn, R. N. et al. Transgenic lambda medaka as a new model for germ cell mutagenesis. Environ. Mol. Mutagen. 49, 173–184 (2008).
Dubrova, Y. E., Plumb, M., Gutierrez, B., Boulton, E. & Jeffreys, A. J. Transgenerational mutation by radiation. Nature 405, 37 (2000).
Dubrova, Y. E. Radiation-induced transgenerational instability. Oncogene 22, 7087–7093 (2003).
Makova, K. D. & Li, W. H. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416, 624–626 (2002).
Miyata, T., Hayashida, H., Kuma, K., Mitsuyasu, K. & Yasunaga, T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 52, 863–867 (1987).
Taylor, J., Tyekucheva, S., Zody, M., Chiaromonte, F. & Makova, K. D. Strong and weak male mutation bias at different sites in the primate genomes: insights from the human–chimpanzee comparison. Mol. Biol. Evol. 23, 565–573 (2006).
Drost, J. B. & Lee, W. R. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ. Mol. Mutagen. 25, 48–64 (1995).
Shen, J. C., Rideout, W. M. 3rd & Jones, P. A. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976 (1994).
Trasler, J. M. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod. Fertil. Dev. 18, 63–69 (2006).
Lees-Murdock, D. J. & Walsh, C. P. DNA methylation reprogramming in the germ line. Epigenetics 3, 5–13 (2008).
Eckhardt, F. et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nature Genet. 38, 1378–1385 (2006).
El-Maarri, O. et al. Methylation levels at selected CpG sites in the factor VIII and FGFR3 genes, in mature female and male germ cells: implications for male- driven evolution. Am. J. Hum. Genet. 63, 1001–1008 (1998).
Morgan, H. D., Dean, W., Coker, H. A., Reik, W. & Petersen-Mahrt, S. K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues. J. Biol. Chem. 279, 52353–52360 (2004).
Schreck, S. et al. Activation-induced cytidine deaminase (AID) is expressed in normal spermatogenesis but only infrequently in testicular germ cell tumours. J. Pathol. 210, 26–31 (2006).
Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA 100, 4102–4107 (2003).
Olsen, A.-K., Lindeman, B., Wiger, R., Duale, N. & Brunborg, G. How do male germ cells handle DNA damage? Toxicol. Appl. Pharmacol. 207 (Suppl. 2), 521–531 (2005).
Jaroudi, S. & SenGupta, S. DNA repair in mammalian embryos. Mutat. Res. 635, 53–77 (2007).
Menezo, Y. Jr, Russo, G., Tosti, E., El Mouatassim, S. & Benkhalifa, M. Expression profile of genes coding for DNA repair in human oocytes using pangenomic microarrays, with a special focus on ROS linked decays. J. Assist. Reprod. Genet. 24, 513–520 (2007).
Intano, G. W. et al. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol. Cell. Biol. 22, 2410–2418 (2002).
Xu, G. et al. Nucleotide excision repair activity varies among murine spermatogenic cell types. Biol. Reprod. 73, 123–130 (2005).
Cortazar, D., Kunz, C., Saito, Y., Steinacher, R. & Schar, P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst.) 6, 489–504 (2007).
Schroering, A. G., Edelbrock, M. A., Richards, T. J. & Williams, K. J. The cell cycle and DNA mismatch repair. Exp. Cell Res. 313, 292–304 (2007).
Hardeland, U., Kunz, C., Focke, F., Szadkowski, M. & Schar, P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 35, 3859–3867 (2007).
Crow, J. F. Spontaneous mutation in man. Mutat. Res. 437, 5–9 (1999).
Risch, N., Reich, E. W., Wishnick, M. M. & McCarthy, J. G. Spontaneous mutation and parental age in humans. Am. J. Hum. Genet. 41, 218–248 (1987).
Ketterling, R. P. et al. Germline origins in the human F9 gene: frequent G:C-->A:T mosaicism and increased mutations with advanced maternal age. Hum. Genet. 105, 629–640 (1999).
Wilkie, A. O. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 16, 187–203 (2005).
Edghill, E. L. et al. Origin of de novo KCNJ11 mutations and risk of neonatal diabetes for subsequent siblings. J. Clin. Endocrinol. Metab. 92, 1773–1777 (2007).
Kubota, H. & Brinster, R. L. Culture of rodent spermatogonial stem cells, male germline stem cells of the postnatal animal. Methods Cell Biol. 86, 59–84 (2008).
McLean, D. J. Spermatogonial stem cell transplantation, testicular function, and restoration of male fertility in mice. Methods Mol. Biol. 450, 149–162 (2008).
Falciatori, I., Lillard-Wetherell, K., Wu, Z., Hamra, F. K. & Garbers, D. L. Deriving mouse spermatogonial stem cell lines. Methods Mol. Biol. 450, 181–192 (2008).
Walsh, C. P. & Xu, G. L. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 301, 283–315 (2006).
Klose, R. J. & Bird, A. P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Acknowledgements
This work was supported in part by grants from the National Institute of General Medical Sciences (N.A. and P.C.) and the Ellison Medical Research Foundation (N.A.).
Author information
Authors and Affiliations
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
Glossary
- Ligase chain reaction
-
Amplification of a small DNA fragment by successive rounds of DNA ligation using one pair of adjacent primers for each of the two complementary target DNA strands.
- Effective population size
-
The number of breeding individuals in an idealized population that would show similar characteristics to the population under consideration. For a number of reasons, the effective population size is typically smaller than the actual number of individuals in the population.
- Achondroplasia
-
A common form of dwarfism, inherited in an autosomal dominant fashion.
- Apert syndrome
-
An autosomal dominant disorder characterized by premature closing of cranial sutures and fused fingers and toes.
- Spermatogonia
-
Premeiotic diploid cells of the mature male germ line.
- Transversion mutation
-
A point mutation in which a purine base is substituted for a pyrimidine base and vice versa; for example, an A˙T to C˙G transversion.
- Multiple endocrine neoplasia type 2B
-
Mutation in the proto-oncogene RET. The mutation is inherited in an autosomal dominant fashion and leads to early childhood thyroid cancer.
- Transition mutation
-
A point mutation in which a purine base (adenine or guanine) is substituted for a different purine base, and a pyrimidine base (cytosine or thymidine) is substituted for a different pyrimidine base; for example, an A˙T to G˙C transition.
- Rett syndrome
-
AnXlinked neurodevelopmental disorder that is associated with mental retardation. It is found sporadically and almost exclusively in females who inherit a new mutation in the methyl-CpG-binding protein 2 gene (MECP2) from their father.
- Transcription-coupled repair
-
A form of DNA repair that removes DNA lesions that inhibit the progression of RNA polymerase during transcription. The repair process specifically targets lesions on the template strand.
- Biased gene conversion
-
A non-reciprocal copy and paste of one allele onto the other one at heterozygous loci during meiotic recombination. Some authors have proposed that this process is biased such that at a site heterozygous for a G˙C or C˙G allele and an A˙T or T˙A allele there will be more G˙C or C˙G gametes produced.
- Recombination fraction
-
Estimate of the proportion of all gametes that were derived from meiotic crossing-over events in a chosen interval.
- Epigenomics
-
Analysis of epigenetic marks (DNA and protein modifications) on a genome-wide scale.
- Duchenne muscular dystrophy
-
A disorder caused by mutations in the X-linked dystrophin gene and characterized by rapidly worsening muscle weakness.
- Haemophilia A
-
A blood clotting disease resulting from mutations in the X-linked factor VIII gene.
- CpG island
-
A region at least several hundred base pairs in length that is characterized by a high GC content and a large number of unmethylated CpG dinucleotides. CpG islands are found to overlap a large fraction of human gene promoters.
- Bisulphite sequencing
-
Chemical treatment of genomic DNA before sequencing that allows identification of those cytosines that were methylated in the DNA from a particular tissue source. Unmethylated cytosines are converted to uracils, whereas methylated cytosines remain unmodified.
Rights and permissions
About this article
Cite this article
Arnheim, N., Calabrese, P. Understanding what determines the frequency and pattern of human germline mutations. Nat Rev Genet 10, 478–488 (2009). https://doi.org/10.1038/nrg2529
Issue Date:
DOI: https://doi.org/10.1038/nrg2529
This article is cited by
-
Novel structures and evolution of tRNA genes: insight into the chloroplast tRNAs of family Sapindaceae
Genetic Resources and Crop Evolution (2024)
-
Paternal impact on the life course development of obesity and type 2 diabetes in the offspring
Diabetologia (2019)
-
Advanced paternal age effects in neurodevelopmental disorders—review of potential underlying mechanisms
Translational Psychiatry (2017)
-
A germline or de novo mutation in two families with Gaucher disease: implications for recessive disorders
European Journal of Human Genetics (2013)
-
Germline selection for deleterious mutation in males
Nature Reviews Genetics (2012)