Key Points
-
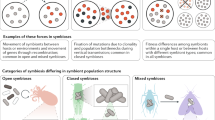
The relevance of symbiotic microbial associations for evolution remained unclear for several decades for theoretical, conceptual, experimental and technological reasons. However, as Darwin emphasized, the struggle for existence includes the dependence of one organism on another.
-
The phylogenetic distribution of symbioses between eukaryotic hosts and prokaryotes is much wider than previously thought, which reinforces the view that this phenomenon has a role in promoting evolutionary innovation. The complete genome sequences for the species that are involved, and the development of metagenomic approaches, provide new opportunities for research into symbioses.
-
Numerous studies indicate that extant endosymbionts originated from ancient pathogenic bacteria that attenuated — or even domesticated — their virulence factors. Adaptation to intracellular life has also involved dramatic genomic and metabolic changes, the most remarkable being genome reduction by gene loss.
-
In many cases, hosts with primary endosymbionts are invaded by new symbionts that, although they are facultative, can provide benefits to the host. These new bacteria could take over the symbiotic functions of the initial symbiont, and so the situation might end in the replacement of one symbiont by the other. Alternatively, a microbial consortium can be established, owing to metabolic complementation between symbionts.
-
The host must also develop mechanisms to adapt its innate immune system, to house the bacteria and to control their proliferation. This is an emerging research area, and its findings provide insights into the non-pathogenicity of endosymbionts. The genome sequencing of selected hosts will be crucial for a complete overview of these associations.
-
The existence of minimal natural genomes has provided important insights for researchers who are interested in experimental genome minimization. Analyses of minimal genomes provide an estimate of the smallest number of genetic elements that are sufficient to build a modern-type free-living cellular organism — a preliminary step in the attempts to make a living cell.
Abstract
Our understanding of prokaryote–eukaryote symbioses as a source of evolutionary innovation has been rapidly increased by the advent of genomics, which has made possible the biological study of uncultivable endosymbionts. Genomics is allowing the dissection of the evolutionary process that starts with host invasion then progresses from facultative to obligate symbiosis and ends with replacement by, or coexistence with, new symbionts. Moreover, genomics has provided important clues on the mechanisms driving the genome-reduction process, the functions that are retained by the endosymbionts, the role of the host, and the factors that might determine whether the association will become parasitic or mutualistic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hypsa, V. & Dale, C. In vitro culture and phylogenetic analysis of 'Candidatus Arsenophonus triatominarum', an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 47, 1140–1144 (1997).
Dale, C. & Maudlin, I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49, 267–275 (1999).
Dale, C., Beeton, M., Harbison, C., Jones, T. & Pontes, M. Isolation, pure culture, and characterization of 'Candidatus Arsenophonus arthropodicus', an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl. Environ. Microbiol. 72, 2997–3004 (2006).
Darby, A. C., Chandler, S. M., Welburn, S. C. & Douglas, A. E. Aphid-symbiotic bacteria cultured in insect cell lines. Appl. Environ. Microbiol. 71, 4833–4839 (2005).
Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 14, 255–274 (1967).
Sapp, J. Evolution by Association. A History of Symbiosis (Oxford University Press, New York, 1994). This book is the first historical report on the symbiosis concept and its central role in shaping contemporary evolutionary ideas about the origin and diversification of life.
de Duve, C. The origin of eukaryotes: a reappraisal. Nature Rev. Genet. 8, 395–403 (2007).
Margulis, L. Symbiosis in Cell Evolution. Microbial Communities in the Archaean and Proterozoic Eons (W. H. Freeman and Co., New York, 1993).
Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (2000).
Tamas, I. et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296, 2376–2379 (2002).
Akman, L. et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nature Genet. 32, 402–407 (2002).
van Ham, R. C. et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl Acad. Sci. USA 100, 581–586 (2003).
Gil, R. et al. The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc. Natl Acad. Sci. USA 100, 9388–9393 (2003).
Degnan, P. H., Lazarus, A. B. & Wernegreen, J. J. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 15, 1023–1033 (2005).
Wu, D. et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4, e188 (2006).
Nakabachi, A. et al. The 160-kilobase genome of the bacterial endosymbiont. Carsonella Science 314, 267 (2006).
Newton, I. L. et al. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315, 998–1000 (2007).
Kuwahara, H. et al. Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr. Biol. 17, 881–886 (2007).
Foster, J. et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3, e121 (2005).
Nakagawa, S. et al. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl Acad. Sci. USA 104, 12146–12150 (2007).
Pérez-Brocal, V. et al. A small microbial genome: the end of a long symbiotic relationship? Science 314, 312–313 (2006). This paper, together with reference 16, shows that the genomes of B. aphidicola BCc and C. ruddii represent the lower limits of genome size in insect endosymbionts. Further research in these systems might shed light on the evolutionary fate of endosymbionts.
Toh, H. et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156 (2006).
McCutcheon, J. P. & Moran, N. A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl Acad. Sci. USA 104, 19392–19397 (2007).
Dale, C. & Moran, N. A. Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465 (2006).
Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189 (2005).
Russell, J. A., Latorre, A., Sabater-Munoz, B., Moya, A. & Moran, N. A. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12, 1061–1075 (2003).
Chen, D.-Q., Montllor, C. B. & Purcell, A. H. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95, 315–323 (2000).
Montllor, C. B., Maxmen, A. & Purcell, A. H. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195 (2002).
Russell, J. A. & Moran, N. A. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. Biol. Sci. 273, 603–610 (2006).
Oliver, K. M., Moran, N. A. & Hunter, M. S. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl Acad. Sci. USA 102, 12795–12800 (2005).
Oliver, K. M., Russell, J. A., Moran, N. A. & Hunter, M. S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl Acad. Sci. USA 100, 1803–1807 (2003).
Scarborough, C. L., Ferrari, J. & Godfray, H. C. Aphid protected from pathogen by endosymbiont. Science 310, 1781 (2005).
Tsuchida, T., Koga, R. & Fukatsu, T. Host plant specialization governed by facultative symbiont. Science 303, 1989 (2004).
Simon, J. C. et al. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. Biol. Sci. 270, 1703–1712 (2003).
Leonardo, T. E. & Muiru, G. T. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc. Biol. Sci. 270 (Suppl 2), S209–S212 (2003).
Leonardo, T. E. & Mondor, E. B. Symbiont modifies host life-history traits that affect gene flow. Proc. Biol. Sci. 273, 1079–1084 (2006).
McGraw, E. A. & O'Neill, S. L. Wolbachia pipientis: intracellular infection and pathogenesis in Drosophila. Curr. Opin. Microbiol. 7, 67–70 (2004).
Welburn, S. C. & Maudlin, I. Tsetse–trypanosome interactions: rites of passage. Parasitol. Today 15, 399–403 (1999).
Moran, N. A. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl Acad. Sci. USA 104 (Suppl 1), 8627–8633 (2007).
Gomez-Valero, L. et al. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 186, 6626–6633 (2004).
Takiya, D. M., Tran, P. L., Dietrich, C. H. & Moran, N. A. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15, 4175–4191 (2006).
Woyke, T. et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955 (2006). This paper and reference 15 are two pioneering works on the application of genomic analysis to uncultivable microbial consortia, which opened up a new field of research on symbiosis.
Giere, O. & Erséus, C. Taxonomy and new bacterial symbioses of gutless marine Tubificidae (Annelida, Oligochaeta) from the island of Elba (Italy). Org. Divers. Evol. 2, 289–297 (2002).
Wernegreen, J. J. Genome evolution in bacterial endosymbionts of insects. Nature Rev. Genet. 3, 850–861 (2002).
Moran, N. A. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA 93, 2873–2878 (1996).
Itoh, T., Martin, W. & Nei, M. Acceleration of genomic evolution caused by enhanced mutation rate in endocellular symbionts. Proc. Natl Acad. Sci. USA 99, 12944–12948 (2002).
Wernegreen, J. J. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr. Opin. Genet. Dev. 15, 572–583 (2005).
Rocha, E. P. & Danchin, A. Base composition bias might result from competition for metabolic resources. Trends Genet. 18, 291–294 (2002).
Fares, M. A., Ruiz-Gonzalez, M. X., Moya, A., Elena, S. F. & Barrio, E. Endosymbiotic bacteria: GroEL buffers against deleterious mutations. Nature 417, 398 (2002).
Moya, A., Latorre, A., Sabater-Munoz, B. & Silva, F. J. Comparative molecular evolution of primary (Buchnera) and secondary symbionts of aphids based on two protein-coding genes. J. Mol. Evol. 55, 127–137 (2002).
Rispe, C., Delmotte, F., van Ham, R. C. & Moya, A. Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res. 14, 44–53 (2004).
Moran, N. A. & Mira, A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2, RESEARCH0054 (2001).
Moran, N. A. & Plague, G. R. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14, 627–633 (2004).
Plague, G. R., Dunbar, H. E., Tran, P. L. & Moran, N. A. Extensive proliferation of transposable elements in heritable bacterial symbionts. J. Bacteriol. 190, 777–779 (2007).
Heddi, A., Charles, H., Khatchadourian, C., Bonnot, G. & Nardon, P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C content of an endocytobiotic DNA. J. Mol. Evol. 47, 52–61 (1998).
Lefevre, C. et al. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21, 965–973 (2004).
Wu, M. et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2, e69 (2004).
Silva, F. J., Latorre, A. & Moya, A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17, 615–618 (2001).
Gomez-Valero, L., Silva, F. J., Simon, J. C. & Latorre, A. Genome reduction of the aphid endosymbiont Buchnera aphidicola in a recent evolutionary time scale. Gene 389, 87–95 (2007).
Gomez-Valero, L., Latorre, A. & Silva, F. J. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 21, 2172–2181 (2004).
Tamames, J. et al. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 7, 181 (2007).
Wilson, A. C. et al. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics 7, 50 (2006).
Wilcox, J. L., Dunbar, H. E., Wolfinger, R. D. & Moran, N. A. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48, 1491–1500 (2003).
Koga, R., Tsuchida, T. & Fukatsu, T. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. Biol. Sci. 270, 2543–2550 (2003).
Ren, Q. & Paulsen, I. T. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1, e27 (2005).
Fiala- Médioni, A. & Métivier, C. Ultrastructure of the gill of the hydrothermal vent bivalve Calyptogena magnifica, with a discussion of its nutrition. Mar. Biol. 90, 215–222 (1986).
Baumann, P., Moran, N. A. & Baumann, L. in The Prokaryotes (ed. Dworkin, M.) 1–67 (Springer, New York, 2000).
Fiala-Médioni, A., Michalski, J. C., Jollès, J., Alonso, C. & Montreuil, J. Lysosomic and lysozyme activities in gills of bivalves from deep hydrothermal vents. C. R. Acad. Sci. Paris 317, 239–244 (1994).
Nakabachi, A. et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl Acad. Sci. USA 102, 5477–5482 (2005).
Dale, C., Young, S. A., Haydon, D. T. & Welburn, S. C. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl Acad. Sci. USA 98, 1883–1888 (2001).
Young, G. M., Schmiel, D. H. & Miller, V. L. A new pathway for the secretion of virulence factors by bacteria: The flagellar export apparatus functions as a protein-secretion system. Proc. Natl Acad. Sci. USA 96, 6456–6461 (1999).
Maezawa, K. et al. Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J. Bacteriol. 188, 6539–6543 (2006).
McFall-Ngai, M. Adaptive immunity: care for the community. Nature 445, 153 (2007).
Hoffmann, J. A. The immune response of Drosophila. Nature 426, 33–38 (2003).
Heddi, A. et al. Molecular and cellular profiles of insect bacteriocytes: mutualism and harm at the initial evolutionary step of symbiogenesis. Cell. Microbiol. 7, 293–305 (2005).
Zaidman-Remy, A. et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473 (2006).
Anselme, C., Vallier, A., Balmand, S., Fauvarque, M.-O. & Heddi, A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl. Environ. Microbiol. 72, 6766–6772 (2006).
Ghedin, E. et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 (2007). The complete genome sequence of the nematode B. malayi , together with the previously reported genome sequence of its Wolbachia endosymbiont, will enable a systems approach for the study of host–symbiont interactions.
Kato, Y. & Komatsu, S. ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum. Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem. 271, 30493–30498 (1996).
Braendle, C. et al. Developmental origin and evolution of bacteriocytes in the aphid–Buchnera symbiosis. PLoS Biol. 1, e21 (2003). This paper was the first study of the development and evolution of a bacteriocyte containing endosymbiotic bacteria.
Lwoff, A. L'évolution physiologique. Étude des pertes des fonctions chez les microorganismes (Hermann, Paris, 1944).
Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev. Genet. 5, 123–135 (2004).
Martin, W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc. Natl Acad. Sci. USA 100, 8612–8614 (2003).
Hotopp, J. C. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007).
Bhattacharya, D., Archibald, J. M., Weber, A. P. M. & Reyes-Prieto, A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29, 1239–1246 (2007).
C. elegans Sequencing Consortium. Genome sequence of the nematode Caenorhabditis elegans: a platform for investigating biology. Science 282, 2012–2018 (1998).
Myers, E. W. et al. A whole-genome assembly of Drosophila. Science 287, 2196–2204 (2000).
Kennedy, J., Marchesi, J. R. & Dobson, A. D. Metagenomic approaches to exploit the biotechnological potential of the microbial consortia of marine sponges. Appl. Microbiol. Biotechnol. 75, 11–20 (2007).
Martin, F. P. et al. A top-down systems biology view of microbiome–mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3, 112 (2007).
Paracer, S. & Ahmadjian, V. Symbiosis. An Introduction to Biological Associations (Oxford University Press, New York, 2000).
Kneip, C., Lockhart, P., Voss, C. & Maier, U. G. Nitrogen fixation in eukaryotes — new models for symbiosis. BMC Evol. Biol. 7, 55 (2007).
Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61, 262–280 (1997).
Stewart, F. J., Newton, I. L. & Cavanaugh, C. M. Chemosynthetic endosymbioses: adaptations to oxic–anoxic interfaces. Trends Microbiol. 13, 439–448 (2005).
Minic, Z. & Herve, G. Biochemical and enzymological aspects of the symbiosis between the deep-sea tubeworm Riftia pachyptila and its bacterial endosymbiont. Eur. J. Biochem. 271, 3093–3102 (2004).
Zientz, E., Dandekar, T. & Gross, R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68, 745–770 (2004).
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J. & Smith, A. G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005).
Buchner, P. Endosymbiosis of Animals with Plant Microorganisms (Interscience, New York, 1965).
Taylor, M. W., Radax, R., Steger, D. & Wagner, M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347 (2007).
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Stingl, U., Radek, R., Yang, H. & Brune, A. 'Endomicrobia': cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71, 1473–1479 (2005).
Fieseler, L., Quaiser, A., Schleper, C. & Hentschel, U. Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ. Microbiol. 8, 612–624 (2006).
Moreira, D. et al. Global eukaryote phylogeny: Combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol. Phylogenet. Evol. 44, 255–266 (2007).
Rodriguez-Ezpeleta, N. et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr. Biol. 17, 1420–1425 (2007).
Lecointre, G. & Le Guyader, H. The Tree of Life. A Phylogenetic Classification (Harvard University Press Reference Library) (Belknap Press, Cambridge, 2006).
Delsuc, F., Brinkmann, H., Chourrout, D. & Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006).
Bourlat, S. J. et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444, 85–88 (2006).
O'Malley, M., Powell, A., Davies, J. & Calvert, J. Knowledge-making distinctions in synthetic biology. Bioessays 30, 57–65 (2007).
Luisi, P. L. Chemical aspects of synthetic biology. Chem. Biodivers. 4, 603–621 (2007).
Peretó, J. & Català, J. The renaissance of synthetic biology. Biol. Theor. 2, 128–130 (2007).
Mushegian, A. R. & Koonin, E. V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA 93, 10268–10273 (1996).
Gil, R., Silva, F. J., Pereto, J. & Moya, A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68, 518–537 (2004). This Review proposes a minimal genetic repertoire for a hypothetical heterotrophic bacterium, based on comparative genomics of endosymbionts, experimentally determined gene essentiality and the functions of a coherent metabolism.
Klasson, L. & Andersson, S. G. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 12, 37–43 (2004).
Gabaldon, T. et al. Structural analyses of a hypothetical minimal metabolism. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 362, 1751–1762 (2007).
Ruiz-Mirazo, K., Pereto, J. & Moreno, A. A universal definition of life: autonomy and open-ended evolution. Orig. Life Evol. Biosph. 34, 323–346 (2004).
Acknowledgements
Financial support was provided by grants GV/2007/050 from Generalitat Valenciana, Spain, to R.G and BFU2006-06003 from Ministerio de Educación y Ciencia (MEC), Spain, to A.L. R.G is a recipient of a contract in the 'Ramón y Cajal' programme from the MEC, Spain. Our thanks to H. Escrivá (CNRS, Banyuls sur Mer), P. López (CNRS, Orsay) and D. Moreira (CNRS, Orsay) for their advice in the design of the eukaryotic phylogenetic tree in BOX 1.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Symbiosis
-
From the Greek, sym 'with' and biosis 'living'. A long-term association between two or more organisms of different species that is integrated at the behavioural, metabolic or genetic level. According to the level of dependence on the host, symbiosis can be obligate or facultative. The term was introduced by Anton de Bary and Albert Bernard Frank when discussing lichens and mycorrhizae, respectively, at the end of the 1870s.
- Parasitism
-
Symbioses in which one species is increasing its fitness while the fitness of the other species is adversely affected.
- Mutualism
-
Symbioses in which both species increase their fitness.
- Commensalism
-
Symbioses in which one partner is increasing its fitness without affecting the other species.
- Ectosymbiosis
-
A symbiosis in which the symbiont lives on the body surface of the host, including internal surfaces such as the lining of the digestive tube and the ducts of glands.
- Endosymbiosis
-
Symbioses in which a prokaryote symbiont lives inside a eukaryotic cell.
- Primary endosymbiont
-
(P-endosymbiont). Obligate bacterial endosymbionts that live inside specialized animal host cells called bacteriocytes. The association is obligate for both partners.
- Secondary symbiont
-
(S-symbiont). Facultative bacterial endosymbiont that coexists with a P-endosymbiont. Often located in syncitial cells near the bacteriocyte and in various other insect tissue types. Secondary symbionts are not essential for host survival and are transferred horizontally among individuals of both the host species and other species.
- Metagenomics
-
The application of genomic analyses to uncultured microorganisms. Also referred to as environmental genomics.
- Vertical transmission
-
The endosymbionts are maternally transferred, that is, directly from a host to its offspring.
- Bacteriocytes
-
Specialized cells of the host species in which symbiotic bacteria live.
- Heterotrophy
-
The metabolic mode in which the carbon source is organic matter. By extension, this is a metabolic mode in which organic matter is the source of carbon, electrons and energy (chemoorganoheterotrophy).
- Chemolithoautotrophy
-
The metabolic mode in which CO2 is the carbon source and an inorganic chemical reaction is both the electron and energy source.
- Horizontal transmission
-
Some endosymbionts retain a generalized ability to colonize and persist in multiple hosts, that is, their transmission is between individuals of the same or different host species, rather than from parent to offspring.
- Syntrophy
-
Emergence of new metabolic capabilities as a result of symbiosis, it is often essential for the survival of the consortium.
- Bacteriome
-
An organ-like structure formed by bacteriocytes.
- Pathogenic island
-
A part of a genome, for which there is evidence of its acquisition by horizontal transfer, that encodes genes that contribute to the virulence of a pathogen.
- Minimal genome
-
The smallest set of genes that is necessary and sufficient to sustain a living cell in the most favourable conditions; that is, in the presence of adequate nutrients and in the absence of stress factors.
- Synthetic biology
-
The design and fabrication of artificial biological systems, with the aim of either optimizing their performance in the context of their technological utility or of deepening our understanding of the naturally occurring organisms.
Rights and permissions
About this article
Cite this article
Moya, A., Peretó, J., Gil, R. et al. Learning how to live together: genomic insights into prokaryote–animal symbioses. Nat Rev Genet 9, 218–229 (2008). https://doi.org/10.1038/nrg2319
Issue Date:
DOI: https://doi.org/10.1038/nrg2319
This article is cited by
-
Genesis of ectosymbiotic features based on commensalistic syntrophy
Scientific Reports (2024)
-
Obligate mutualistic heritable symbiosis in sap-feeding insects: an intricate relationship in nature
Symbiosis (2024)
-
Coordination of host and endosymbiont gene expression governs endosymbiont growth and elimination in the cereal weevil Sitophilus spp.
Microbiome (2023)
-
Host-Adapted Strains of Spodoptera frugiperda Hold and Share a Core Microbial Community Across the Western Hemisphere
Microbial Ecology (2023)
-
Synthase-selected sorting approach identifies a beta-lactone synthase in a nudibranch symbiotic bacterium
Microbiome (2023)