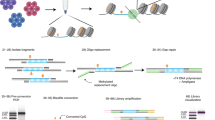
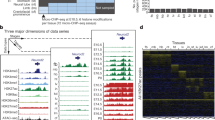
Key Points
-
Chromatin modifications have been shown to have a profound impact on the regulation of gene expression.
-
Epigenomes consist of the ensemble of all chromatin modifications in any given cell type, including DNA methylation, post-translational histone modifications, nucleosome positioning, histone variants, noncoding RNAs and three-dimensional chromatin architecture.
-
New technologies, which allow for the profiling of chromatin modifications on a genome-wide scale, are providing researchers with comprehensive views of epigenomes.
-
Genome-scale data sets for epigenetic phenomena allow for the use of bioinformatic methods to study epigenetics.
-
Different functional regions of the genome are associated with distinct patterns of histone modifications and these patterns, in turn, can be used to annotate the functional elements in the genome.
Abstract
Over two metres of DNA is packaged into each nucleus in the human body in a manner that still allows for gene regulation. This remarkable feat is accomplished by the wrapping of DNA around histone proteins in repeating units of nucleosomes to form a structure known as chromatin. This chromatin structure is subject to various modifications that have profound influences on gene expression. Recently developed techniques to study chromatin modifications at a genome-wide scale are now allowing researchers to probe the complex components that make up epigenomes. Here we review genome-wide approaches to studying epigenomic structure and the exciting findings that have been obtained using these technologies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Goldberg, A. D., Allis, C. D. & Bernstein, E. Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007).
Ptashne, M. On the use of the word 'epigenetic'. Curr. Biol. 17, R233–R236 (2007).
Bird, A. Perceptions of epigenetics. Nature 447, 396–398 (2007).
Bernstein, B. E., Meissner, A. & Lander, E. S. The mammalian epigenome. Cell 128, 669–681 (2007).
Orlando, V. & Paro, R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75, 1187–1198 (1993).
Blat, Y. & Kleckner, N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98, 249–259 (1999).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000). This paper introduced the ChIP–chip technique, used here to map Gal4 and Ste12 binding sites in the yeast genome.
Bernstein, B. E. et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA 99, 8695–8700 (2002).
Robyr, D. et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109, 437–446 (2002).
Robyr, D. & Grunstein, M. Genomewide histone acetylation microarrays. Methods 31, 83–89 (2003).
Bernstein, B. E., Liu, C. L., Humphrey, E. L., Perlstein, E. O. & Schreiber, S. L. Global nucleosome occupancy in yeast. Genome Biol. 5, R62 (2004).
Lee, C. K., Shibata, Y., Rao, B., Strahl, B. D. & Lieb, J. D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 36, 900–905 (2004).
Ozsolak, F., Song, J. S., Liu, X. S. & Fisher, D. E. High-throughput mapping of the chromatin structure of human promoters. Nature Biotechnol. 25, 244–248 (2007). This study mapped nucleosome positions across ∼3,700 promoters in seven human cell lines using MNase digestion followed by hybridization to tiling microarrays.
Impey, S. et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041–1054 (2004).
Roh, T. Y., Ngau, W. C., Cui, K., Landsman, D. & Zhao, K. High-resolution genome-wide mapping of histone modifications. Nature Biotechnol. 22, 1013–1016 (2004).
Wei, C. L. et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 (2006).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein–DNA interactions. Science 316, 1497–1502 (2007).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods 4, 651–657 (2007).
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). Together with Reference 17, these studies were the first to demonstrate how ChIP–Seq can be used to profile histone modifications and DNA-binding sites across the entire human genome.
Feinberg, A. P. Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–440 (2007).
Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev. Genet. 8, 286–298 (2007).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Gardiner-Garden, M. & Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282 (1987).
Takai, D. & Jones, P. A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA 99, 3740–3745 (2002).
Ng, H. H. & Bird, A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9, 158–163 (1999).
Ioshikhes, I. P. & Zhang, M. Q. Large-scale human promoter mapping using CpG islands. Nature Genet. 26, 61–63 (2000).
Bell, A. C., West, A. G. & Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 (1999).
Hark, A. T. et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000).
Tate, P. H. & Bird, A. P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 3, 226–231 (1993).
Robertson, K. D. & Wolffe, A. P. DNA methylation in health and disease. Nature Rev. Genet. 1, 11–19 (2000).
Baylin, S. B. & Herman, J. G. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16, 168–174 (2000).
Jones, P. A. & Laird, P. W. Cancer epigenetics comes of age. Nature Genet. 21, 163–167 (1999).
Zilberman, D. & Henikoff, S. Genome-wide analysis of DNA methylation patterns. Development 134, 3959–3965 (2007).
Bird, A. P. & Southern, E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol. 118, 27–47 (1978).
Selker, E. U. et al. The methylated component of the Neurospora crassa genome. Nature 422, 893–897 (2003).
Khulan, B. et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 16, 1046–1055 (2006).
Lippman, Z. et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 (2004).
Yan, P. S. et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 61, 8375–8380 (2001).
Hatada, I. et al. A microarray-based method for detecting methylated loci. J. Hum. Genet. 47, 448–451 (2002).
Rollins, R. A. et al. Large-scale structure of genomic methylation patterns. Genome Res. 16, 157–163 (2006).
Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA 89, 1827–1831 (1992).
Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA 93, 9821–9826 (1996).
Eads, C. A. et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 28, e32 (2000).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Adorjan, P. et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 30, e21 (2002).
Gitan, R. S., Shi, H., Chen, C. M., Yan, P. S. & Huang, T. H. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 12, 158–164 (2002).
Dupont, J. M., Tost, J., Jammes, H. & Gut, I. G. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 333, 119–127 (2004).
Mockler, T. C. et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics 85, 1–15 (2005).
Bibikova, M. et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 16, 383–393 (2006).
Rakyan, V. K. et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2, e405 (2004).
Eckhardt, F. et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nature Genet. 38, 1378–1385 (2006). The follow-up study from the Human Epigenome Project consortium, which profiled DNA methylation on three human chromosomes for several healthy tissues and primary cells by sequencing bisulphite-treated DNA.
Keshet, I. et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nature Genet. 38, 149–153 (2006).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genet. 37, 853–862 (2005).
Zhang, X. et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201 (2006). The first comprehensive map of DNA methylation for an entire genome, produced by performing mCIP combined with tiling microarrays with 35 bp resolution.
Zilberman, D., Gehring, M., Tran, R. K., Ballinger, T. & Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genet. 39, 61–69 (2007).
Shen, L. et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 3, 2023–2036 (2007).
Weber, M. et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 39, 457–466 (2007).
Das, R. et al. Computational prediction of methylation status in human genomic sequences. Proc. Natl Acad. Sci. USA 103, 10713–10716 (2006).
Fang, F., Fan, S., Zhang, X. & Zhang, M. Q. Predicting methylation status of CpG islands in the human brain. Bioinformatics 22, 2204–2209 (2006).
Bock, C. et al. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2, e26 (2006).
Bock, C., Walter, J., Paulsen, M. & Lengauer, T. CpG island mapping by epigenome prediction. PLoS Comput. Biol. 3, e110 (2007).
Roth, S. Y. & Allis, C. D. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17, 93–98 (1992).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Turner, B. M. Histone acetylation and an epigenetic code. Bioessays 22, 836–845 (2000).
Schreiber, S. L. & Bernstein, B. E. Signaling network model of chromatin. Cell 111, 771–778 (2002).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Schubeler, D. et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 (2004).
Liu, C. L. et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3, e328 (2005).
Pokholok, D. K. et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 (2005).
Bernstein, B. E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Koch, C. M. et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 17, 691–707 (2007).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007).
Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005).
Roh, T. Y., Cuddapah, S., Cui, K. & Zhao, K. The genomic landscape of histone modifications in human T cells. Proc. Natl Acad. Sci. USA 103, 15782–15787 (2006).
Roh, T. Y., Cuddapah, S. & Zhao, K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552 (2005).
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007).
Ng, H. H., Robert, F., Young, R. A. & Struhl, K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 (2003).
Martens, J. H. et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812 (2005).
Boyer, L. A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Squazzo, S. L. et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16, 890–900 (2006).
Stock, J. K. et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature Cell Biol. 9, 1428–1435 (2007).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Roh, T. Y., Wei, G., Farrell, C. M. & Zhao, K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 17, 74–81 (2007).
Meneghini, M. D., Wu, M. & Madhani, H. D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 (2003).
Raisner, R. M. et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 (2005).
Zhang, H., Roberts, D. N. & Cairns, B. R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231 (2005).
Guillemette, B. et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384 (2005).
Li, B. et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA 102, 18385–18390 (2005).
Guillemette, B. & Gaudreau, L. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 84, 528–535 (2006).
Mito, Y., Henikoff, J. G. & Henikoff, S. Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411 (2007).
Mito, Y., Henikoff, J. G. & Henikoff, S. Genome-scale profiling of histone H3.3 replacement patterns. Nature Genet. 37, 1090–1097 (2005).
Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct. Biol. 7, 1121–1124 (2000).
Jin, C. & Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529 (2007).
Lohr, D. & Lopez, J. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1–10 and GAL80 genes. J. Biol. Chem. 270, 27671–27678 (1995).
Straka, C. & Horz, W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 10, 361–368 (1991).
Yuan, G. C. et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 (2005). This study profiled nucleosome positions at high resolution across most of chromosome 3 of the S. cerevisiae genome with MNase digestion followed by hybridization to DNA microarrays.
Lee, W. et al. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genet. 39, 1235–1244 (2007).
Johnson, S. M., Tan, F. J., McCullough, H. L., Riordan, D. P. & Fire, A. Z. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 16, 1505–1516 (2006).
Albert, I. et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 (2007).
Barski, A. et al. Response: mapping nucleosome positions using ChIP-Seq data. Cell 131, 832–833 (2007).
Schmid, C. D. & Bucher, P. ChIP–Seq data reveal nucleosome architecture of human promoters. Cell 131, 831–832 (2007).
Satchwell, S. C., Drew, H. R. & Travers, A. A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191, 659–675 (1986).
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006).
Ioshikhes, I. P., Albert, I., Zanton, S. J. & Pugh, B. F. Nucleosome positions predicted through comparative genomics. Nature Genet. 38, 1210–1215 (2006).
Peckham, H. E. et al. Nucleosome positioning signals in genomic DNA. Genome Res. 17, 1170–1177 (2007).
Gilbert, N. et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118, 555–566 (2004).
Crawford, G. E. et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl Acad. Sci. USA 101, 992–997 (2004).
Weil, M. R., Widlak, P., Minna, J. D. & Garner, H. R. Global survey of chromatin accessibility using DNA microarrays. Genome Res. 14, 1374–1381 (2004).
Crawford, G. E. et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nature Methods 3, 503–509 (2006).
Crawford, G. E. et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 (2006).
Xi, H. et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136 (2007).
Nagy, P. L., Cleary, M. L., Brown, P. O. & Lieb, J. D. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl Acad. Sci. USA 100, 6364–6369 (2003).
Hogan, G. J., Lee, C. K. & Lieb, J. D. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2, e158 (2006).
Giresi, P. G., Kim, J., McDaniell, R. M., Iyer, V. R. & Lieb, J. D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Liu, X. S. Getting started in tiling microarray analysis. PLoS Comput. Biol. 3, 1842–1844 (2007).
Ji, H. & Wong, W. H. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics 21, 3629–3636 (2005).
Johnson, W. E. et al. Model-based analysis of tiling-arrays for ChIP–chip. Proc. Natl Acad. Sci. USA 103, 12457–12462 (2006).
Marinescu, V. D. et al. START: an automated tool for serial analysis of chromatin occupancy data. Bioinformatics 22, 999–1001 (2006).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nature Genet. 39, 1522–1527 (2007).
Spilianakis, C. G. & Flavell, R. A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nature Immunol. 5, 1017–1027 (2004).
Spilianakis, C. G., Lalioti, M. D., Town, T., Lee, G. R. & Flavell, R. A. Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645 (2005).
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G. & Cremer, T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8, 104–115 (2007).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002). This paper introduced the chromosome conformation capture (3C) technique.
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genet. 38, 1341–1347 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature Genet. 38, 1348–1354 (2006).
Dostie, J. et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Hall, I. M. et al. Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002).
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Grewal, S. I. & Elgin, S. C. Transcription and RNA interference in the formation of heterochromatin. Nature 447, 399–406 (2007).
Cam, H. P. et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genet. 37, 809–819 (2005).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Kim, T. H. & Ren, B. Genome-wide analysis of protein–DNA interactions. Annu. Rev. Genomics Hum. Genet. 7, 81–102 (2006).
van Steensel, B. & Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nature Biotechnol. 18, 424–428 (2000).
Eissenberg, J. C. & Elgin, S. C. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210 (2000).
Tamaru, H. & Selker, E. U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414, 277–283 (2001).
Vire, E. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2006).
O'Neill, L. P., VerMilyea, M. D. & Turner, B. M. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nature Genet. 38, 835–841 (2006).
Dahl, J. A. & Collas, P. Q2C hIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells 25, 1037–1046 (2007).
Wolffe, A. P. & Hayes, J. J. Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720 (1999).
Acknowledgements
We thank members of the Zhao laboratory for helpful discussions. We apologize to those whose work was not included here owing to space limitations. Research in the authors' laboratory is supported by the Intramural Research Program of the US National Institutes of Health, National Heart, Lung and Blood Institute.
Author information
Authors and Affiliations
Related links
Related links
FURTHER INFORMATION
Glossary
- Chromatin immunoprecipitation
-
A technique to isolate individual chromatin fragments using an antibody that is specific to a feature of the chromatin fragments (for example, a DNA-binding protein, a particular histone modification, or DNA methylation).
- ChIP–chip
-
The combination of ChIP experiments with DNA microarrays to profile protein targeting or chromatin modifications over large genomic regions.
- Serial analysis of gene expression
-
A sequence-based quantitative technique that is used to determine mRNA levels. cDNA is generated from an mRNA sample, digested with a four-base cutter and ligated to an adaptor containing a class II restriction enzyme that releases a 14 to 21 bp fragment. The short fragments are concatenated together, cloned into a sequencing vector and sequenced.
- ChIP–Seq
-
The combination of ChIP experiments with high-throughput sequencing to quantitatively analyse protein targeting or chromatin modifications across the entire genome.
- Tiling microarrays
-
DNA microarrays with densely spaced or overlapping probes that allow for high-resolution genomic mapping.
- Fluorescence in situ hybridization
-
A technique that involves the fluorescent labelling of single-stranded DNA probes that then target specific regions of chromosomes and allow for the visualization of these regions within the cell.
- 3C
-
Chromosome conformation capture. A technique that is used to study the long-distance interactions between genomic regions, which in turn can be used to study the three-dimensional architecture of chromosomes within a cell nucleus.
- 4C
-
Either chromosome conformation capture-on-chip or circular chromosome conformation capture. These techniques allow the profiling of many interactions throughout a genome with a specific locus.
- 5C
-
Chromosome conformation capture carbon copy. A high-throughput extension of 3C that pairs the 3C technology with DNA microarrays or high-throughput sequencing. This technique allows the profiling of many chromatin interactions in parallel.
Rights and permissions
About this article
Cite this article
Schones, D., Zhao, K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet 9, 179–191 (2008). https://doi.org/10.1038/nrg2270
Issue Date:
DOI: https://doi.org/10.1038/nrg2270
This article is cited by
-
New genetic and epigenetic insights into the chemokine system: the latest discoveries aiding progression toward precision medicine
Cellular & Molecular Immunology (2023)
-
Interactions of free-living amoebae with the rice fungal pathogen, Rhizoctonia solani
BMC Research Notes (2019)
-
Integrated analysis of DNA-methylation and gene expression using high-dimensional penalized regression: a cohort study on bone mineral density in postmenopausal women
BMC Medical Genomics (2018)
-
Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells
Nature Reviews Genetics (2017)
-
Epigenetic Regulation of Sost/sclerostin Expression
Current Molecular Biology Reports (2017)