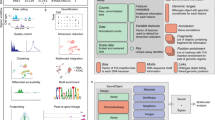
Key Points
-
Chromatin is a highly dynamic complex, and chromatin dynamics operate over several orders of magnitude of space and time.
-
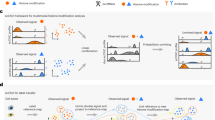
Local chromatin dynamics are involved in dictating access for transcription factors (TFs), the gene expression machinery and other chromatin effectors. Local chromatin dynamics can be probed in vitro and in vivo using a range of experimental approaches, including single-molecule force spectroscopy and single-molecule tracking methods.
-
The long-range establishment, maintenance and remodelling of chromatin states in the 3D space of the nucleus are involved in the regulation of transcriptional programmes and cell differentiation.
-
Genome-wide studies based on time-resolved chromatin immunoprecipitation and single-cell omics enable the interrogation of dynamic processes on the genomic scale and the single-cell scale.
-
For a complete picture of chromatin function, short-range and rapid dynamics need to be integrated with slower dynamics on the genomic scale; such integration will require new experimental and theoretical approaches.
Abstract
The establishment and maintenance of chromatin states involves multiscale dynamic processes integrating transcription factor and multiprotein effector dynamics, cycles of chemical chromatin modifications, and chromatin structural organization. Recent developments in genomic technologies are emerging that are enabling a view beyond ensemble- and time-averaged properties and are revealing the importance of dynamic chromatin states for cell fate decisions, differentiation and reprogramming at the single-cell level. Concurrently, biochemical and single-molecule methodologies are providing key insights into the underlying molecular mechanisms. Combining results from defined in vitro and single-molecule studies with single-cell genomic approaches thus holds great promise for understanding chromatin-based transcriptional memory and cell fate. In this Review, we discuss recent developments in biochemical, single-molecule biophysical and single-cell genomic technologies and review how the findings from these approaches can be integrated to paint a comprehensive picture of dynamic chromatin states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cacchiarelli, D. et al. Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell 162, 412–424 (2015).
Henikoff, S. Mechanisms of nucleosome dynamics in vivo. Cold Spring Harb. Perspect. Med. 6, a026666 (2016).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Takahashi, K. & Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183–193 (2016).
Heard, E. & Martienssen, R. A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 (2014).
Fierz, B. & Muir, T. W. Chromatin as an expansive canvas for chemical biology. Nat. Chem. Biol. 8, 417–427 (2012).
Yu, J. Single-molecule studies in live cells. Annu. Rev. Phys. Chem. 67, 565–585 (2016).
Schwartzman, O. & Tanay, A. Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet. 16, 716–726 (2015).
Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015).
Hubner, M. R. & Spector, D. L. Chromatin dynamics. Annu. Rev. Biophys. 39, 471–489 (2010).
Soutoglou, E. & Misteli, T. Mobility and immobility of chromatin in transcription and genome stability. Curr. Opin. Genet. Dev. 17, 435–442 (2007).
Luger, K., Dechassa, M. L. & Tremethick, D. J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13, 436–447 (2012).
Casolari, J. M. et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439 (2004).
Osborne, C. S. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36, 1065–1071 (2004).
Akhtar, A. & Gasser, S. M. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8, 507–517 (2007).
Vazquez, J., Belmont, A. S. & Sedat, J. W. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11, 1227–1239 (2001).
Tumbar, T. & Belmont, A. S. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3, 134–139 (2001).
Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186 (2001).
Chubb, J. R., Boyle, S., Perry, P. & Bickmore, W. A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Choy, J. S. & Lee, T. H. Structural dynamics of nucleosomes at single-molecule resolution. Trends Biochem. Sci. 37, 425–435 (2012).
Li, G., Levitus, M., Bustamante, C. & Widom, J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 (2005).
Kruithof, M. et al. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 16, 534–540 (2009).
Poirier, M. G., Oh, E., Tims, H. S. & Widom, J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 16, 938–944 (2009).
Li, W. et al. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol. Cell 64, 120–133 (2016). This paper provides insights into the internal energetics and dynamics of chromatin arrays by force spectroscopy, and how nucleosome–nucleosome interactions are modulated by a protein factor.
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Azzaz, A. M. et al. Human heterochromatin protein 1alpha promotes nucleosome associations that drive chromatin condensation. J. Biol. Chem. 289, 6850–6861 (2014).
Hiragami-Hamada, K. et al. Dynamic and flexible H3K9me3 bridging via HP1beta dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 7, 11310 (2016).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Talbert, P. B. & Henikoff, S. Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 (2010).
Dekker, J. & Misteli, T. Long-range chromatin interactions. Cold Spring Harb. Perspect. Biol. 7, a019356 (2015).
Teves, S. S., Weber, C. M. & Henikoff, S. Transcribing through the nucleosome. Trends Biochem. Sci. 39, 577–586 (2014).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Fierz, B. Dynamic chromatin regulation from a single molecule perspective. ACS Chem. Biol. 11, 609–620 (2015).
Polach, K. J. & Widom, J. Mechanism of protein access to specific DNA-sequences in chromatin — a dynamic equilibrium-model for gene-regulation. J. Mol. Biol. 254, 130–149 (1995).
Li, G. & Widom, J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11, 763–769 (2004).
Chien, F. T. & van Noort, J. 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr. Pharm. Biotechnol. 10, 474–485 (2009).
Poirier, M. G., Bussiek, M., Langowski, J. & Widom, J. Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 379, 772–786 (2008).
Moyle-Heyrman, G., Tims, H. S. & Widom, J. Structural constraints in collaborative competition of transcription factors against the nucleosome. J. Mol. Biol. 412, 634–646 (2011).
Hieb, A. R., Gansen, A., Bohm, V. & Langowski, J. The conformational state of the nucleosome entry–exit site modulates TATA box-specific TBP binding. Nucleic Acids Res. 42, 7561–7576 (2014).
Tims, H. S., Gurunathan, K., Levitus, M. & Widom, J. Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 411, 430–448 (2011).
Taylor, I. C., Workman, J. L., Schuetz, T. J. & Kingston, R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 5, 1285–1298 (1991).
Polach, K. J. & Widom, J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258, 800–812 (1996).
Mirny, L. A. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA 107, 22534–22539 (2010).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Koopmans, W. J., Brehm, A., Logie, C., Schmidt, T. & van Noort, J. Single-pair FRET microscopy reveals mononucleosome dynamics. J. Fluoresc. 17, 785–795 (2007).
Wei, S., Falk, S. J., Black, B. E. & Lee, T. H. A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic Acids Res. 43, e111 (2015).
Bohm, V. et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 39, 3093–3102 (2011).
Ngo, T. T. & Ha, T. Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 43, 3964–3971 (2015).
Sinha, K. K., Gross, J. D. & Narlikar, G. J. Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 355, eaaa3761 (2017).
Bucceri, A., Kapitza, K. & Thoma, F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 25, 3123–3132 (2006).
Neuman, K. C. & Nagy, A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008).
Rudnizky, S. et al. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 7, 12958 (2016).
Hall, M. A. et al. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16, 124–129 (2009).
Ngo, T. T., Zhang, Q., Zhou, R., Yodh, J. G. & Ha, T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 160, 1135–1144 (2015). This study uses a combination of optical tweezers and single-molecule FRET to demonstrate that nucleosome-unwrapping dynamics depend on the local DNA sequence.
Jonkers, I. & Lis, J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015).
Deindl, S. & Zhuang, X. Monitoring conformational dynamics with single-molecule fluorescence energy transfer: applications in nucleosome remodeling. Methods Enzymol. 513, 59–86 (2012).
Narlikar, G. J., Sundaramoorthy, R. & Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013).
Bartholomew, B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 83, 671–696 (2014).
Venkatesh, S. & Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Schalch, T., Duda, S., Sargent, D. F. & Richmond, T. J. X-Ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141 (2005).
Song, F. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 (2014).
Hsieh, T. H. et al. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 162, 108–119 (2015).
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Elgin, S. C. & Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5, a017780 (2013).
Muller, K. P. et al. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys. J. 97, 2876–2885 (2009).
Cheutin, T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003).
Festenstein, R. et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299, 719–721 (2003).
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Ayoub, N., Jeyasekharan, A. D., Bernal, J. A. & Venkitaraman, A. R. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453, 682–686 (2008).
Kilic, S., Bachmann, A. L., Bryan, L. C. & Fierz, B. Multivalency governs HP1alpha association dynamics with the silent chromatin state. Nat. Commun. 6, 7313 (2015). This study uses single-molecule imaging in a highly defined chromatin system to monitor effector protein dynamics in real time as a function of the local chromatin state and effector multivalency.
Tanay, A. & Cavalli, G. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr. Opin. Genet. Dev. 23, 197–203 (2013).
Boettiger, A. N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
McNally, J. G., Muller, W. G., Walker, D., Wolford, R. & Hager, G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008).
Gebhardt, J. C. et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013).
Chen, J. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014).
Manley, S. et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008).
Normanno, D. et al. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 6, 7357 (2015).
Liu, Z., Lavis, L. D. & Betzig, E. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 58, 644–659 (2015).
Coleman, R. A. et al. Imaging transcription: past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 80, 1–8 (2015).
Elf, J., Li, G. W. & Xie, X. S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 (2007).
Izeddin, I. et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3, e02230 (2014).
Morisaki, T., Muller, W. G., Golob, N., Mazza, D. & McNally, J. G. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 5, 4456 (2014).
Swinstead, E. E. et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 (2016).
Li, M. et al. Dynamic regulation of transcription factors by nucleosome remodeling. eLife 4, e06249 (2015).
Blackledge, N. P., Rose, N. R. & Klose, R. J. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16, 643–649 (2015).
Zhen, C. Y. et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. eLife 5, e17667 (2016). Chromatin effector dynamics are monitored in real-time in live cells using SMT methods, demonstrating the collaboration between DNA binding and histone PTM recognition in the molecular targeting of effectors on chromatin.
Fegan, A., White, B., Carlson, J. & Wagner, C. Chemically controlled protein assembly: techniques and applications. Chem. Rev. 110, 3315–3336 (2010).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012). In this paper, chemically induced dimerization is used in an elegant approach to monitor heterochromatin spreading and inheritance in live cells.
Hodges, C. & Crabtree, G. Dynamics of inherently bounded histone modification domains. Proc. Natl Acad. Sci. USA 109, 13296–13301 (2012).
Bintu, L. et al. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724 (2016).
Schones, D. E. & Zhao, K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 9, 179–191 (2008).
Zhang, Z. & Pugh, B. F. High-resolution genome-wide mapping of the primary structure of chromatin. Cell 144, 175–186 (2011).
Zentner, G. E. & Henikoff, S. High-resolution digital profiling of the epigenome. Nat. Rev. Genet. 15, 814–827 (2014).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Ram, O. et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147, 1628–1639 (2011).
Liang, J. et al. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol. Cell 53, 672–681 (2014).
Kasinathan, S., Orsi, G. A., Zentner, G. E., Ahmad, K. & Henikoff, S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods 11, 203–209 (2014).
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016).
Lesne, A., Riposo, J., Roger, P., Cournac, A. & Mozziconacci, J. 3D genome reconstruction from chromosomal contacts. Nat. Methods 11, 1141–1143 (2014).
Ulianov, S. V. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 26, 70–84 (2016).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Nagano, T. et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 10, 1986–2003 (2015).
Andrey, G. et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013).
Battich, N., Stoeger, T. & Pelkmans, L. Control of transcript variability in single mammalian cells. Cell 163, 1596–1610 (2015).
Rhee, H. S. & Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
Rhee, H. S., Bataille, A. R., Zhang, L. & Pugh, B. F. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 159, 1377–1388 (2014).
Neph, S. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012).
Sung, M. H., Guertin, M. J., Baek, S. & Hager, G. L. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell 56, 275–285 (2014).
He, H. H. et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 11, 73–78 (2014).
Gusmao, E. G., Allhoff, M., Zenke, M. & Costa, I. G. Analysis of computational footprinting methods for DNase sequencing experiments. Nat. Methods 13, 303–309 (2016).
Lickwar, C. R., Mueller, F., Hanlon, S. E., McNally, J. G. & Lieb, J. D. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484, 251–255 (2012).
Poorey, K. et al. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science 342, 369–372 (2013). This study establishes CLK-ChIP to monitor TF dynamics from seconds to minutes at several genomic loci, demonstrating large-scale variability in residence times that are modulated by cofactors.
van Steensel, B., Delrow, J. & Henikoff, S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27, 304–308 (2001).
van Steensel, B. & Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000).
Aughey, G. N. & Southall, T. D. Dam it's good! DamID profiling of protein–DNA interactions. Wiley Interdiscip. Rev. Dev. Biol. 5, 25–37 (2016).
Wang, H., Mayhew, D., Chen, X., Johnston, M. & Mitra, R. D. Calling cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res. 21, 748–755 (2011).
Zentner, G. E., Kasinathan, S., Xin, B., Rohs, R. & Henikoff, S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 6, 8733 (2015). In this paper, ChEC-seq is used as a time-sensitive and high-throughput method to profile TF binding dynamics genome-wide, allowing the discrimination of different locus-specific TF binding modes.
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Schmid, M. et al. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21, 379–391 (2006).
Halligan, D. L. & Keightley, P. D. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40, 151–172 (2009).
Shipony, Z. et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 513, 115–119 (2014). An analysis of the methylome in clonal populations of pluripotent and somatic cells reveals that pluripotent cells rapidly turn over their epigenetic signatures, resulting in a stable epigenetic state, in contrast to somatic cells in which epimutations are inherited.
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011).
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 (2011).
Audergon, P. N. et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015).
Ragunathan, K., Jih, G. & Moazed, D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015). In references 127 and 128, rapid turnover or epigenetic inheritance through transmission of H3K9 methylation is demonstrated in fission yeast, as a function of the efficiency of counteracting mechanisms by a specific demethylase.
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Ramskold, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Head, S. R. et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 56, 61–64 (2014).
Shalek, A. K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014).
Wen, L. & Tang, F. Single-cell sequencing in stem cell biology. Genome Biol. 17, 71 (2016).
van Galen, P. et al. A multiplexed system for quantitative comparisons of chromatin landscapes. Mol. Cell 61, 170–180 (2016).
Shema, E. et al. Single-molecule decoding of combinatorially modified nucleosomes. Science 352, 717–721 (2016).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Voigt, P. et al. Asymmetrically modified nucleosomes. Cell 151, 181–193 (2012).
Weiner, A. et al. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat. Biotechnol. 34, 953–961 (2016).
Lechner, C. C., Agashe, N. D. & Fierz, B. Traceless synthesis of asymmetrically modified bivalent nucleosomes. Angew. Chem. Int. Ed. 55, 2903–2906 (2016).
Kim, D.-H. et al. Histone H3 K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol. Cell. Biol. 33, 4936–4946 (2013).
Hu, D. et al. Not all H3K4 methylations are created equal: Mll2/COMPASS dependency in primordial germ cell specification. Mol. Cell 65, 460–475.e6 (2017).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013).
Macaulay, I. C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Dey, S. S., Kester, L., Spanjaard, B., Bienko, M. & van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 33, 285–289 (2015).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Cheow, L. F. et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13, 833–836 (2016).
Wu, C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286, 854–860 (1980).
Jin, W. et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015).
Auerbach, R. K. et al. Mapping accessible chromatin regions using Sono-Seq. Proc. Natl Acad. Sci. USA 106, 14926–14931 (2009).
Giresi, P. G., Kim, J., McDaniell, R. M., Iyer, V. R. & Lieb, J. D. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Gaulton, K. J. et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 (2010).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015)
Ramachandran, S. & Henikoff, S. Transcriptional regulators compete with nucleosomes post-replication. Cell 165, 580–592 (2016). In this paper, MINCE-seq is presented as a new method to directly assess the chromatin landscape at replication forks with high temporal and spatial resolution, demonstrating that epigenetic information needs be re-established after replication.
Jaitin, D. A. et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell 167, 1883–1896.e15 (2016).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Battich, N., Stoeger, T. & Pelkmans, L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 10, 1127–1133 (2013).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Coskun, A. F. & Cai, L. Dense transcript profiling in single cells by image correlation decoding. Nat. Methods 13, 657–660 (2016).
Filion, G. J. et al. Systematic protein location mapping reveals five principal chromatin types in drosophila cells. Cell 143, 212–224 (2010).
Kharchenko, P. V. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011).
Roy, S. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006).
Musselman, C., Lalonde, M.-E., Côté, J. & Kutateladze, T. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 (2004).
Thiru, A. et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23, 489–499 (2004).
Ryu, H. W. et al. Analysis of the heterochromatin protein 1 (HP1) interactome in Drosophila. J. Proteomics 102, 137–147 (2014).
Talbert, P. B. & Henikoff, S. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 7, 793–803 (2006).
Al-Sady, B., Madhani, H. & Narlikar, G. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol. Cell 51, 80–91 (2013).
Muller, M. M., Fierz, B., Bittova, L., Liszczak, G. & Muir, T. W. A two-state activation mechanism controls the histone methyltransferase Suv39h1. Nat. Chem. Biol. 12, 188–193 (2016).
Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Gaydos, L. J., Rechtsteiner, A., Egelhofer, T. A., Carroll, C. R. & Strome, S. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2, 1169–1177 (2012).
Lhoumaud, P. et al. Insulators recruit histone methyltransferase dMes4 to regulate chromatin of flanking genes. EMBO J. 33, 1599–1613 (2014).
Canzio, D. et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 (2011).
Li, P. L. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Margueron, R. & Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010).
Acknowledgements
Dedicated to the memory of Jörg Langowski. B.F. gratefully acknowledges the Sandoz Family Foundation, the Swiss National Science Foundation (grant 31003A_149789), the NCCR Chemical Biology, Systems X (51PHP0_163580) and EPFL for financial support. O.C. acknowledges CNRS/INSB and the grant support of Fondation pour la Recherche Médicale (Equipe FRM DEQ20160334940).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Transcriptional programmes
-
A subset of genes actively transcribed in a subpopulation of cells, a cell type or at a developmental stage. Transcriptional programmes depend on transcription factors, chromatin modifications and dynamics to regulate transcription initiation, elongation and termination.
- Local feedback loops
-
Networks of effector activity that result in the amplification (positive feedback) or the dampening (negative feedback) of a local perturbation in the system.
- Chromatin effector proteins
-
Proteins or protein complexes that interact with and alter the local function of chromatin; for example, by modulating chromatin structure, chemically modifying DNA or histones, or by recruiting further biochemical activity to this site.
- Emergent properties
-
Properties of a complex molecular system that are not observable in the individual molecules but that emerge from their interactions.
- Multivalent interactions
-
The simultaneous engagement of multiple binding sites between two binding partners; for example, the simultaneous binding of multiple histone post-translational modifications by an effector protein that contains several specific reader domains, resulting in an increase in binding affinity.
- Chromatin-remodelling complexes
-
Protein complexes that use ATP to move nucleosomes, to exchange histone variants, or to disrupt and remove nucleosomes, thereby contributing to the regulation of chromatin structure, composition and gene regulation.
- Fluorescence correlation spectroscopy
-
(FCS). A spectroscopic method that allows the determination of dynamic parameters (such as molecular diffusion and structural dynamics) from the time-correlation of intensity fluctuations of the sample fluorescence emission.
- Fluorescence recovery after photobleaching
-
(FRAP). A microscopic method to measure the diffusion and interaction kinetics of protein factors in cells. Fluorescently labelled proteins are bleached in the confocal microscope at a defined site, followed by measurement of the local recovery of fluorescence by back-diffusion of fluorescent molecules.
- Single-molecule tracking
-
(SMT). A method to track single, fluorescently labelled (or otherwise tagged) molecules, particularly in living cells, using microscopy techniques. Computational analysis of the individual tracks yields local residence times, diffusion coefficients and diffusion behaviour, depending on the cellular environment.
- Chromatin immunoprecipitation
-
(ChIP). Affinity enrichment of formaldehyde-fixed, fragmented (usually by sonication) chromatin using an antibody against an epitope of interest, such as a histone post-translational modification or a bound protein, followed by DNA identification using PCR amplification or sequencing.
- Pioneer TFs
-
Transcription factors (TFs) that initially bind to the regulatory regions of silenced genes, resulting in opening of the chromatin structure, the facilitation of the assembly of further factors, and thus leading to gene activation.
- Fluorescence resonance energy transfer
-
(FRET). A non-radiative, distance-dependent energy transfer process between a donor and an acceptor fluorophore. It enables the measurement of the physical distance between the dye pairs as a function of transfer efficiency. When dyes are at defined sites within macromolecules, structural transitions can be measured in real time.
- Force spectroscopy
-
Methods to study interaction forces between molecules by manipulating the interaction partners using mechanical means, magnetism or light.
- Optical tweezers
-
A force spectroscopy method in which microscopic beads (to which the molecules under investigation are attached) are trapped within the focal spot of a laser. Modulating the strength or the position of the optical trap then exerts defined forces onto the target molecules.
- Nucleosome dyad
-
The central axis of the nucleosome, dividing the particle into two halves related by pseudo two-fold symmetry.
- Total internal reflection fluorescence
-
(TIRF). A microscopic method in which the sample is illuminated by an evanescent field that decays within a few hundred nanometres distance from the coverslip. This allows the imaging of molecules close to the surface with very low background, making this method highly useful for single-molecule imaging.
- Confocal single-molecule detection
-
A microscopy technique that focuses the excitation light onto a diffraction-limited spot and removes out-of-focus emission light by the addition of a pinhole at the confocal plane of the imaging lens. Only molecules contained in the confocal volume (in the order of femtolitres) are observed, making this method useful for single-molecule observation.
- Cryo-electron-microscopy
-
(Cryo-EM). A transmission electron microscopy technique of unstained samples in a vitrified water layer at cryogenic temperature. Particle averaging of homogenous sample preparation allows 3D reconstruction of the molecular structure, making cryo-EM a highly powerful tool for structural biology.
- Micrococcal nuclease
-
(MNase). A highly active endo-exonuclease that rapidly digests all DNA that is not protected by bound proteins, thus it is often used for protein footprinting applications.
- Enhanceosome
-
A set of factors forming a higher-order protein complex that can recognize enhancer DNA, followed by transcription activation through interactions with core transcriptional factors.
- Paired-end sequencing
-
A DNA sequencing method in which each individual DNA fragment is sequenced from both the 5′ and the 3′ ends, thus increasing sequencing fidelity and facilitating the detection of genomic rearrangements and mapping of repetitive regions.
- Affinity enrichment
-
Enrichment of a molecule from a complex mixture based on its affinity to a specific binder, for example, an antibody.
- Chromosome conformation capture (3C) techniques
-
Analysis methods of genome organization by crosslinking DNA and chromatin-interacting proteins in situ within the nucleus, followed by DNA fragmentation, extraction of the crosslinked products and DNA re-ligation. Analysis of the DNA ligation products produces maps indicating regions of contact within the nucleus, thus allowing a reconstruction of the native chromatin structure. Several 3C-derived techniques exist: 3C investigates interactions between a pair of loci, 4C interrogates interactions between a single locus and the whole genome, 5C captures all interactions within a defined genomic locus of limited size, and Hi-C probes the interactions between all genomic loci.
- Epimutation accumulation approach
-
Monitors the stability of existing epigenomic signatures and the emergence of alternative patterns, including DNA methylation, histone modifications and gene expression traits, in clonal cell populations over time.
- Bisulfite sequencing
-
DNA methylation profiling through DNA sequencing upon bisulfite treatment, which converts cytosine into uracil but leaves 5-methylcytosine (5mC) unaffected. Comparison to the original sequence reveals 5mC sites.
- Unique molecular identifiers
-
(UMIs). Barcoding of DNA molecules by ligation of unique sequence identifiers for the absolute counting of independent molecules in a sequencing library. This allows DNA molecules obtained independently (distinct UMIs) to be distinguished from sequences that originate from the same fragment (identical UMI), for example, due to PCR-mediated pre-amplification steps in library preparation.
Rights and permissions
About this article
Cite this article
Cuvier, O., Fierz, B. Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat Rev Genet 18, 457–472 (2017). https://doi.org/10.1038/nrg.2017.28
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2017.28
This article is cited by
-
Alteration of chromosome structure impacts gene expressions implicated in pancreatic ductal adenocarcinoma cells
BMC Genomics (2024)
-
Interrogating epigenetic mechanisms with chemically customized chromatin
Nature Reviews Genetics (2024)
-
CSCS: a chromatin state interface for Chinese Spring bread wheat
aBIOTECH (2021)
-
Insulator-based loops mediate the spreading of H3K27me3 over distant micro-domains repressing euchromatin genes
Genome Biology (2020)
-
Pollutants inducing epigenetic changes and diseases
Environmental Chemistry Letters (2020)