Key Points
-
Hepatitis C virus (HCV) infection is an emerging global epidemic. The development of therapeutic and preventive strategies is a formidable challenge for biotechnology and pharmaceutical companies owing to the absence of adequate animal models and tissue-culture systems for viral propagation and reconstitution of the virus life cycle and pathogenesis. Despite these obstacles, remarkable progress has been made through collaborative efforts between basic researchers, clinicians and the pharmaceutical indutry.
-
Interferon-α (IFN-α)-based therapies remain the treatment of choice for hepatitis C. To improve the efficacy and safety of IFN therapies, some companies are focusing on modification of the IFN-α molecule, chemically or genetically, to develop long-lasting IFN molecules, whereas others are investigating combination therapies with known and new antivirals. However, most patients do not respond well to the existing IFN-based therapies, and alternative therapeutic interventions are desperately needed for these patients.
-
Several advances in the development of anti-HCV therapeutics have come about owing to: the definition of molecular clones that are infectious in the chimpanzee animal model of HCV infection; the availability of the three-dimensional structures of several of the key virally encoded enzymes; and the recent development of HCV-replicon systems in Huh-7 cells.
-
In vitro assays have been developed to examine viral enzymatic activities for the testing and development of antiviral agents, primarily targeting specific processes that are essential to HCV replication: virus translation; processing of the viral polyprotein protein; and viral RNA replication.
-
Inhibitors of the HCV internal ribosome-entry site (IRES), non-structural protein 3 (NS3) protease and NS5B polymerase are in clinical development at present. Several small-molecule compounds, which inhibit viral replication, and a few prophylactic vaccines, are under investigation for hepatitis C.
-
Cellular genes, pathways or processes required for HCV replication represent an attractive class of genes for small-molecule targeting to control viral replication. Therapeutic strategies that are being developed focus on depleting intracellular pools of guanine nucleotides by inhibiting the cellular IMPDH. Other potential targets include disruption of the host-cell glycosylation machinery for viral morphogenesis and secretion through inhibition of endoplasmic reticulum (ER) α-glucosidases and association of HCV NS proteins with the ER membranes and/or host-cell proteins.
-
Several therapies that preserve the cell structure, or prevent or reverse fibrosis and cirrhosis that are caused by chronic hepatitis C, or prevent the recurrence of hepatitis C after liver-transplant surgery are also under clinical development.
Abstract
Chronic infection with hepatitis C virus (HCV) is an emerging global epidemic. The development of effective HCV antiviral therapeutics continues to be a daunting challenge owing to the absence of adequate animal models and tissue-culture systems for analysis and propagation of the virus. Despite these obstacles, inhibitors of the replicative elements of HCV, immune modulators and non-specific hepatoprotective agents are being pursued and exciting progress has been made. Successful therapeutic intervention of HCV will probably require combination approaches and new approaches, including host drug discovery targets.
Similar content being viewed by others
Main
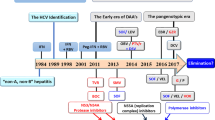
Although hepatitis C virus (HCV) was first identified in 1989 (Ref. 1), its threat as a serious public health problem and disease burden has become recognized only in the past few years. The blood-borne HCV infection is often subclinical, despite persistent and progressive inflammation and FIBROSIS of the liver, which ultimately results in liver CIRRHOSIS, hepatic failure or HEPATOCELLULAR CARCINOMA2. It is believed that the virus has been spread unknowingly since the early 1960s, mainly through blood transfusions, before reliable routine blood tests became available in 1992. More recent cases, however, are associated with intravenous drug use and other unrecognizable transmission factors/routes3. It is estimated that HCV has infected more than 170 million people globally, nearly fives times more than human immunodeficiency virus (HIV)-infected individuals4. This problem is magnified by the high frequency of HCV persistence during infection — the virus establishes a chronic infection in up to 85% of cases2. HCV infection has become the most common cause of hepatocellular carcinoma and the primary reason for liver transplantations among adults in western countries5. So, it is no surprise that the virus has commanded significant attention from academic and pharmaceutical-industry laboratories.
The current therapy for hepatitis C is inadequate at treating all patients who are afflicted with chronic hepatitis C and, so far, there are no broadly effective anti-HCV compounds. It is clear that new and better therapeutic strategies are desperately needed in the battle against HCV. In the past few years, targeted antiviral drugs, such as protease and reverse-transcriptase inhibitors, have had an impressive effect on HIV-related morbidity and mortality. Similarities in the HIV and HCV key enzymes (with respect to conserved subdomain organization and arrangement of structural motifs) indicate that analogous drugs might also be useful in treating hepatitis C (reviewed in Ref. 6). However, there are considerable barriers to the development of anti-HCV therapeutics, which include the persistence of the virus, the genetic diversity during replication in the host, the development of drug-resistant virus mutants and the lack of reproducible infectious culture systems and small-animal models for HCV replication and pathogenesis. Moreover, given the mild clinical course of HCV infection in most cases and the ever-complex biology of the liver, careful consideration must be given to antiviral drugs, which are likely to have marked side effects. Multiple-agent treatment modalities are likely to be needed, probably on a rotation basis, and novel viral and host targets are being explored, which will challenge us to rethink the way we design and carry out clinical trials.
Although combination antiviral therapies will be required to limit the genetic diversity of HCV, an appropriate immuno-based therapy might also be necessary to eliminate completely VIRUS LOAD from the blood. Immunological factors that are likely to be involved in the clearance of HCV after acute infection include CYTOTOXIC T LYMPHOCYTE (CTL) activity, antibody responses, robust T-helper type 1 (TH1)-CELL cytokine involvement and limited TH2-CELL cytokine participation7. Although the CTL response in chronically HCV-infected patients seems strong, it fails to contain the virus infection and therefore allows the smouldering inflammation due to the targeted immune response to HCV to set the stage for hepatocarcinogenesis. The lack of a strong TH1 response is believed to contribute to the chronicity of hepatitis C, and maintaining or inducing a TH1-mediated response might therefore be an important key to HCV treatment. This concept is supported by the clinical use of INTERFERON-α (IFN-α), a cytokine that not only acts through its antiviral effect, but also leads to an enhancement of the TH1 response in HCV treatment.
Many biotechnology and pharmaceutical companies are devoting a substantial level of resources to the discovery and development of new drugs for HCV therapeutic intervention. Approaches vary from targeting established viral targets, to developing protective and/or therapeutic vaccines against HCV, to improving current IFN-based therapies. These have been the subject of several reviews8,9,10,11,12,13. There is also tremendous interest from academia, as indicated by the exponential increase in the number of publications related to hepatitis C in recent years. Equipped with new advances in our understanding of the HCV replication cycle and key enzymes, and the recent availability of HCV-REPLICON systems in Huh-7 cells14 and small-animal models15,16, we are now in a better position to identify and develop anti-HCV agents. Here, we review the current status and emerging strategies for anti-HCV therapeutics. Because most of our current antiviral efforts are aimed at key viral enzymes that are essential to HCV replication, we begin our treatise with a succinct overview of the molecular biology and life cycle of HCV.
Know your foe: HCV genome and life cycle
HCV is a small (40–60 nM in diameter), ENVELOPED, single-stranded (ss) RNA Hepacivirus in the Flaviviridae family; the most closely related human viruses are GB virus C (GBV-C) or hepatitis H virus (HGV), yellow-fever virus and dengue virus17. The HCV genome and its encoded gene products have been characterized in detail (for review, see Ref. 18). The positive-strand RNA genome comprises a 5′ non-translated region (NTR), a single open reading frame (ORF) of ∼9,000 nucleotides in length and a short 3′NTR (Fig. 1). The 5′NTR (∼340 nucleotides) contains the internal ribosome-entry site (IRES), which mediates the initiation of viral-RNA translation in a cap-independent manner. Sequences in the 5′NTR, including the IRES, are essential for replication of this virus19. The 3′NTR (230 nucleotides) comprises a tripartite structure that consists of a variable region immediately after the stop codon of the ORF, a poly (U/UC) tract that varies in length between 30 and 150 residues and a highly conserved 'X-tail' or 3′X sequence. The 3′X region is crucial for efficient RNA replication20.
The boxed area corresponds to the single open reading frame of the hepatitis C virus (HCV) genome. The stem–loop structures represent the 5′ and 3′ non-translated (NTR) regions, including the internal ribosome-entry site (IRES) and 3′X regions. The function and molecular mass (in kDa) of the gene products after polyprotein processing are shown. Core (C)–E1, E1–E2, E2–p7 and p7–non-structural protein 2 (NS2) junctions are cleaved by a cellular signal peptidase(s) to yield structural proteins. The NS2–NS3 metalloproteinase undergoes autocatalytic cleavage, which releases the mature NS3 serine protease. NS3 cleaves the remainder of the NS polypeptide. The two regions that have extreme sequence variability in E2, known as hypervariable regions 1 and 2 (HVR1 and HVR2), are indicated. A region in NS5A, known as the interferon (IFN)-sensitivity-determining region (ISDR), has been linked to the response to IFN-α therapy in some strains of HCV. Both NS5A and E2 have been implicated as antagonists of IFN (for review, see Ref. 34). ARFP/F, alternative reading-frame protein/frameshift protein; LDLR, low-density lipoprotein receptor; RdRp, RNA-dependent RNA polymerase.
The ORF encodes a single polyprotein of ∼3,010 amino acids, which is cleaved co- and post-translationally at several sites by host signal peptidases and HCV-encoded proteases to produce at least three structural proteins (core, E1 and E2), and six non-structural (NS) proteins (NS2, NS3, NS4A NS4B, NS5A and NS5B)21 (Fig. 1). The core protein interacts with viral RNA and forms the NUCLEOCAPSID. E1 and E2 are heavily glycosylated viral-envelope proteins that can interact with plasma membranes of hepatocytes and other cells. E1 and E2 can form heteromeric complexes, although it is not clear whether their association is necessary for binding to cell membranes. Possible roles for the small, hydrophobic protein p7, which is produced by further cleavage between E2 and NS2, include that of an ion channel, or it might have a role in the subcellular localization of VIRION components and assembly of virus particles22 (see below). Recently, another protein of unknown function, which is encoded by an alternative ORF that overlaps the core-protein gene, has been identified. It is known as ARFP (for alternative-reading-frame protein) or F (for frameshift protein)23,24.
The NS proteins encode enzymes or accessory factors that catalyse and regulate the replication of the HCV RNA genome. Processing of the HCV NS polypeptide is catalysed by two virally encoded proteases (Fig. 1). The NS2–NS3 zinc-dependent metalloproteinase undergoes autocatalytic cleavage to produce NS2 and NS3 (Ref. 21). The released NS3 serine protease catalyses the cleavage of the remaining NS polyprotein to yield NS4A, NS4B, NS5A and NS5B, which, together with NS3 and possibly cellular proteins, form the replication complex or 'replicase' of HCV. The carboxy-terminal segment of the NS3 protein also has NUCLEOSIDE TRIPHOSPHATASE (NTPase) and RNA HELICASE activity. The NS4A protein seems to have at least two functions: to form a stable complex with NS3 to facilitate the membrane localization of the NS3–NS4A complex in the endoplasmic reticulum (ER), and to act as a cofactor for NS3 protease activity. NS4B is a relatively hydrophobic 27-kDa protein of unknown function, and might have a role in the modulation of NS5A hyperphosphorylation. The NS5A phosphoprotein is implicated in mediating HCV resistance to IFN therapy. NS5A might also function to regulate viral replication through its interaction with the NS5B protein25, which is the RNA-dependent RNA polymerase (RdRp) that catalyses the replication of HCV RNA.
On the basis of analogies to the closely related flavi- and pestiviruses and our current understanding of recombinant HCV proteins, a model for HCV life cycle has been proposed26 (Fig. 2). HCV infection begins with attachment, which is mediated by a specific interaction between cell-surface molecules on the target cells and the viral-envelope proteins. The mechanisms of virus entry remain unknown, and the receptors for HCV entry are not known at present. It has been proposed that the binding of the E2 glycoprotein to CD81, a tetraspanin molecule that is expressed on hepatocytes and B lymphocytes27, functions to attach HCV to the surface of cells, which allows a subsequent interaction with a more specific entry receptor, such as the low-density lipoprotein (LDL) receptor28,29. Next, the virus is probably engulfed by receptor-mediated ENDOCYTOSIS. The positive-strand HCV genome is then delivered to the cytoplasm, where the RNA is translated, and the polyprotein is processed in the ER. The replicase is assembled at the ER membrane, where it directs the synthesis of intermediate NEGATIVE-STRAND RNA, which is subsequently used as a template for the generation of positive-strand RNAs. The positive-strand RNA is encapsidated with the structural proteins and is presumably enveloped by budding into the lumen of the ER. The packaging of the HCV RNA genome into newly synthesized virions is probably mediated by specific interactions between sequences within the 5′NTR of the HCV genome and the core protein. Finally, infectious virions are thought to be released by transport through the Golgi compartment.
The life cycle of the hepatitis C virus (HCV) has several specific steps, many of which are targets for antiviral drugs: a | attachment; b | endocytosis; c | virion– membrane fusion; d | uncoating; e | translation and polyprotein processing; f | replicase assembly; g | RNA replication; h | viral assembly and ER budding; i | vesicle transport and glycoprotein maturation; j | vesicle fusion and virion release. ER, endoplasmic reticulum; IRES, internal ribosome-entry site; LDLR, low-density lipoprotein receptor; NS, non-structural protein; siRNA, small interfering RNA.
Because of the high replication rate30 and the lack of proof-reading function of NS5B, the HCV genome has high genetic variability. The genomic-sequence variance is not distributed evenly over the genome, with both the 5′NTR and parts of the 3′NTR being the most highly conserved regions. HCV is classified into six main genotypes, which diverge by ∼30% at the nucleotide-sequence level, and there are more than 30 subtypes throughout the world31. Subtypes 1a and 1b are predominant in the United States and Europe, whereas subtype 1b is frequently found in most Asian countries. Importantly, individuals who are infected with genotype 1 are generally more refractory to IFN-α therapy than individuals who are infected with genotypes 2 and 3 (Ref. 32). For some strains of HCV 1b, this is apparently influenced by variability in a region of the NS5A gene that is known as the IFN-sensitivity-determining region or ISDR (Fig. 1; for review, see Ref. 33). Although the exact mechanisms by which HCV counteracts IFN are not fully understood and are often controversial, both the NS5A protein and E2 glycoprotein have been implicated through their ability to inhibit the IFN-induced protein kinase (PKR)34. In addition, HCV exists in individual patients as QUASISPECIES, which differ mainly in the hypervariable regions (HVR1 and -2) of the E2 gene — (Fig. 1). The variability of HCV quasispecies seems to correlate with the clinical outcome of the HCV infection (reviewed in Ref. 35).
HCV infects only humans and chimpanzees, primarily targeting hepatocytes. The determinants of this restricted host and tissue specificity are not understood, and several reports indicate that HCV can also infect other organs and cell types, in particular lymphoid cells36. This extrahepatic infection might contribute to the immune-mediated pathogenesis of chronic liver disease and/or the development of autoimmune diseases, including MIXED CRYOGLOBULINAEMIA and GLOMERULONEPHRITIS. Despite the extremely robust in vivo replication rate of HCV30, efforts to propagate this virus in cell culture have been frustratingly unsuccessful. For now, we have to use the recently generated HCV-replicon system, in which expression of the HCV NS proteins drives the replication of a subgenomic HCV RNA (for review, see Ref. 14 and the article by Ralf Bartenschlager on p911 of this issue). The HCV-replicon RNA replicates to fairly high levels in Huh-7 cells, and provides — for the first time — a genetic system to study HCV replication and a cell-based assay to screen for HCV inhibitors. Propagation of HCV in chimeric mouse models has recently been achieved15,16. In the first model, immunocompromised mice were engrafted with human hepatocytes isolated from fresh livers, and were shown to be susceptible to HCV infection and replication15. In addition, the Trimera system involves the infection of human liver fragments with HCV ex vivo before being transplanted into immunocompromised mice16. Both systems should help to expedite anti-HCV drug evaluation.
IFN-based therapies: the past, present and future
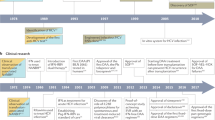
In the absence of an effective antiviral or vaccination strategy against HCV, the single drug that is used to treat chronic HCV infection is IFN-α, a naturally occurring glycoprotein that has antiviral and immunomodulatory properties. It continues to be the only known drug to induce sustained HCV clearance and cause an improvement in liver histology37. However, IFN-α monotherapy is limited by adverse side effects, such as severe flu-like syndrome, LEUKOPAENIA and THROMBOCYTOPAENIA. Furthermore, a SUSTAINED VIROLOGICAL RESPONSE (SVR) is achieved in only 15% of patients, in particular those infected with the most prevalent HCV genotype 1 virus who are carrying high virus loads32.
The combination of the orally active synthetic guanosine analogue ribavirin (Fig. 3) with IFN-α2b (Table 1) has proved to be more effective than IFN-α monotherapy, yielding an SVR in 35–40% of patients38. Ribavirin action is thought to reside, at least in part, in its ability to inhibit inosine monophosphate dehydrogenase (IMPDH), an enzyme that catalyses a rate-limiting step in GTP biosynthesis. This leads to a decreased intracellular pool of GTP levels, and therefore indirectly suppresses the synthesis of viral RNA. The antiviral activity of ribavirin might also be related to its ability to inhibit the HCV NS5B polymerase directly, suppress the humoral and cellular immune responses and/or promote LETHAL MUTAGENESIS of the viral genome (reviewed in Ref. 39). However, despite the improved efficacy of the combination therapy of IFN-α and ribavirin, most patients still fail to achieve an SVR to the treatment32. Furthermore, other side effects have been described for the combination therapy, including HAEMOLYTIC ANAEMIA due to the accumulation of ribavirin triphosphate in erythrocytes38.
To further improve the therapeutic efficacy of IFN-α monotherapy and combination therapy, several options are being investigated. Several studies of the effect of IFN-α treatment on viral kinetics indicated that daily dosing of IFN-α and lengthened treatment duration might improve the SVR30,40. Other therapeutic agents, alone or in combination with IFN-α, have been evaluated. Some clinical benefits were observed in pilot studies with ofloxacin41,42, although these results require confirmation in larger studies. Furthermore, encouraging results have been seen with the immunomodulatory peptide α1-thymosin43 when used in combination with IFN-α, and Phase III hepatitis C clinical trials are underway at present (Table 1). In recent small studies, a triple combination of IFN-α, ribavirin and amantadine was found to improve the SVR44,45.
The best clinical results so far have been achieved with the introduction of polyethylene glycol (PEG)-modified IFN-α, which is characterized by a long half-life, allowing a decreased frequency (weekly) of administration46. Two PEGYLATED IFN-α preparations combined with ribavirin are now standard treatment: Pegasys, a PEGylated IFN-α2a (Fig. 4); and PEG-INTRON, a PEGylated IFN-α2b (Table 1). Treatment with PEGylated IFN-α at least doubles the SVR rates that are achieved with IFN-α monotherapy, or approximates those achieved with IFN-α–ribavirin combination therapy47,48,49. Importantly, combination therapy of PEGylated IFN-α with lower doses of ribavirin reportedly improves the response rate to more than 50% (∼40% in patients infected with HCV genotype 1 and ∼80% in patients carrying genotypes 2 or 3), with fewer serious adverse events, which will probably make this the reigning standard of therapy for chronic hepatitis C. After the success of modified IFN, Phase III trials evaluating Ceplene (histamine dihydrochloride; a histamine analogue) and Pegasys as a combination therapy for HCV treatment are in progress (Table 1). A different modified IFN, in the form of a fusion of ALBUMIN and IFN-α2b (Albuferon-α), is now in Phase I studies in patients for whom conventional IFN treatment for hepatitis C has failed.
The figure depicts a branched polyethylene glycol (PEG) that was created by coupling a monofunctional PEG (mPEG)-benzatriazole carbonate of molecular mass 40 kDa to lysine. Conjugation of this PEG moiety to interferon-α2a (IFN-α2a) results in an agent with a significantly longer half-life, which requires less frequent administration and has an improved toxicity profile. NHS, N-hydroxysuccinimide.
Others have evaluated the use of different or genetically modified IFNs for anti-HCV therapy, including IFN-β50,51, IFN-γ52 and 'consensus' IFN53 (Table 1). Although many of these results need to be reproduced in larger studies, the bioengineered consensus IFN, known as Infergen, which consists of the most frequently observed amino acid at each position of the IFN-α subtypes, is producing encouraging results. Interim data from a Phase IV clinical trial indicated that 56% of patients treated with the combination use of Infergen and ribavirin achieved an SVR. Phase II clinical trials are also underway to evaluate a recombinant human IFN-ω and IFN-β1a in IFN-naive patients with chronic hepatitis (Table 1). Omniferon, which is a naturally occurring IFN-α, is now in Phase II clinical trials in Europe for hepatitis C. Recently, an emerging powerful technology known as MOLECULAR BREEDING or DNA shuffling was used to generate IFN mutants or variants with increased antiviral and antiproliferative activities54. In the shuffling experiment of a family of more than 20 IFN-α genes, the most active human IFN variant was improved 285,000-fold relative to human IFN-α2a. The implications of this approach could be profound in the development of more potent and less toxic IFN molecules, and it would be of great interest to determine whether these shuffled IFNs were active in the HCV-replicon assay.
HCV molecular targets for drug development
Although substantial progress has been achieved with PEGylated IFN-α plus ribavirin therapy, it is evident that most patients with chronic HCV infection are not candidates for IFN-based therapies. Furthermore, IFN-α has limited efficacy in immunocompromised patients, and treatment of HCV/HIV co-infection presents yet another challenge55. So, the development of alternative therapeutic interventions for these patients is imperative56. In this regard, a number of advances have come about, owing to: the definition of molecular clones that are infectious in the chimpanzee animal model of HCV infection; the development of HCV-replicon systems in Huh-7 cells; and the availability of the three-dimensional structures of several of the key virally encoded enzymes, in some cases in complex with substrates, cofactors and inhibitors. In vitro assays have been developed to examine viral enzymatic activities for the testing and development of antiviral agents. These have been aimed primarily at specific processes that are essential to HCV replication, which include: virus translation, which is controlled by regulatory elements, such as the 5′NTR that contains the IRES; processing of the viral protein by the NS2–NS3 and NS3–NS4A proteases; viral RNA replication, which uses the NS3 helicase and NS5B RdRp (Fig. 2).
NS2–NS3 autoprotease. The metal-dependent cysteine protease NS2–NS3 catalyses cleavage between NS2 and NS3 in an autoproteolytic manner57. The amino-terminal portion of NS2 is responsible for membrane association, whereas its carboxy terminus, which overlaps with NS3, is believed to catalyse the cleavage of the NS2–NS3 site. However, the NS2–NS3 proteolytic activity is distinct from the NS3 serine-protease activity. Interestingly, peptides that resemble the NS2–NS3 cleavage site did not inhibit the NS2–NS3 protease activity, whereas cleavage-product-derived peptides and peptides derived from NS4A were found to inactivate NS2–NS3 activity with inhibition constant (Ki) values as low as 3 μM (Refs 58,59). Mutagenesis studies have shown that amino-acid residues His952 and Cys992 are essential for autocatalytic activity60,61. In vitro assays have been developed for the characterization of this enzyme and the identification of new antiviral agents59,62.
NS3 protease. The activity of the chymotrypsin-like serine protease that is encoded within the amino-terminal 180 amino acids of NS3 is indispensable for HCV infectivity in the chimpanzee model63. The structure of the protease domain (Fig. 5) and the full-length NS3 protein were solved by X-ray crystallography64. Efficient processing requires the NS3 protease in combination with the NS4A cofactor and a structural zinc molecule57. Direct comparison of the NS3-protease crystal structure with or without NS4A showed that NS4A is required to improve the anchoring and orientation of the catalytic triad within the serine protease64. In light of the many studies on expression, purification and in vitro enzymatic reconstitution, the NS3 protease is perhaps the most thoroughly characterized HCV enzyme and the most intensively pursued HCV target.
Ribbon conformation of the three-dimensional crystal structures of two hepatitis C virus (HCV) non-structural protein 3 (NS3) serine-protease domains complexed with an NS4A cofactor peptide (residues 956–967; red) and the tripeptide inhibitor (inhibitor I; ball-and-stick model) L-BOC, L-Glu,L-Leu-(difluoro)aminobutyric acid. The zinc ion (indicated in light grey) is present in the non-inhibited structure (bottom) but absent in the inhibited structure (top). BOC, tert-butyloxycarbonyl group.
The NS3 protease is prone to inhibition by specific penta- or hexapeptides derived from the amino-terminal NS3 cleavage products, which have provided the basis for lead optimization of PEPTIDOMIMETIC inhibitors10,65,66. This class of optimized compounds has shown submicromolar potencies in in vitro enzymatic assays, as well as in the HCV-replicon system. Subsequent STRUCTURE–ACTIVITY RELATIONSHIP studies have produced a potent and specific modified tripeptide carboxylic acid (exemplified by the macrocyclic inhibitor in Fig. 6). Other series of structurally diverse NS3-protease substrate-based inhibitors have been disclosed, including α-ketoamides, borinic acids, phosphonates, hydrazinourea, α-ketoacids10 and pyrrolidine-5,5-trans lactams67. Novel chemical scaffolds were also generated using structure-based computational and combinatorial-chemistry techniques, which have resulted in the identification of the lead compound VX-950 (LY-570310). Using a different approach, other groups generated RNA APTAMERS to NS3 that inhibited the protease activity and might, therefore, provide potential leads for molecules that inhibit NS3 or indicate other sites in the NS3 protease for drug intervention68,69.
This compound is a modified tripeptide carboxylic-acid inhibitor of the hepatitis C virus (HCV) non-structural protein 3 (NS3) serine protease. For further details, refer to Ref. 123.
NS3 helicase. The carboxy-terminal 450 amino acids of NS3 define the NTPase/rna helicase domain, the activity of which is also indispensable for viral infectivity in the chimpanzee model63. The HCV helicase has unusual characteristics because it can unwind DNA–RNA and DNA–DNA substrates as well as RNA–RNA hybrids. NS3 helicase activity requires substrates with a 3′ single-stranded overhang, as it unwinds only in a 3′-to-5′ direction57. The structure of the HCV helicase domain, both with and without a single-stranded oligonucleotide, has been determined70,71 (Fig. 7), which might aid in the design of antiviral agents targeted to various sites to inhibit helicase activity. Prospective targets include the NTP-binding site, the binding sites for the ssRNA and double-stranded (ds) RNA or DNA, and the interaction surface between these domains. Oligomerization seems to be necessary for helicase activity and could be an alternative target for intervention72. However, the NS3 helicase has proved to be an extremely challenging target, with little progress being made in the identification of helicase inhibitors compared with inhibitors targeting other HCV proteins.
Ribbon diagram of the hepatitis C virus (HCV) non-structural protein 3 (NS3) helicase RNA-helicase domain complexed with a deoxyuridine octamer (dU8; yellow). The protein comprises three domains. Domains 1 (purple) and 2 (red) share a similar fold, and make symmetrically equivalent contacts with the backbone of the bound oligonucleotide. Domain 3 (green) is predominantly α-helical, and has a carboxy-terminal 40-amino-acid region that lacks any secondary-structure element. A sulphate ion (shown in green) is bound within a phosphate-binding loop in the amino-terminal region of domain 1. Reproduced from Ref. 124 © (1998), with permission from Elsevier Science.
NS4A cofactor: more than a just sidekick? It has been shown that a direct interaction between NS3 and the NS4A cofactor is important for the proteolytic activity of NS3 (Ref. 57), and possibly the helicase activity as well73. Compounds that disrupt or affect the outcome of the protein–protein interaction between NS3 and NS4A (for example, by induction of a conformational change) could potentially interfere with viral polyprotein processing by the NS3 protease. Indeed, several peptides derived from NS4A, which compete with NS4A for binding to NS3, inhibited the protease activity in vitro in low-micromolar values9. NS4A binding also seems to mediate NS3 association with the ER membrane and affect the stability of the NS3 protein74, or internal cleavage of NS3 (Ref. 75) and NS2–NS3 processing58. So, given the multiple functional outcomes assigned to the NS4A–NS3 interaction, it might be an attractive candidate for therapeutic intervention by inhibition of the protein–protein interaction.
NS5B polymerase. HCV NS5B polymerase is also a validated HCV target for antiviral therapy in that its activity is essential for HCV viral replication and infectivity in a chimpanzee model63,76. The biochemical properties of NS5B have been characterized extensively (reviewed in Ref. 77). A detailed view of HCV NS5B was revealed by the crystal structures of the RdRp78,79. Although canonical polymerase features exist, HCV NS5B adopts a unique molecular structure that resembles a 'thumb–palm–finger' that is different from other known DNA and RNA polymerases, highlighting the attractiveness of the HCV polymerase as a drug target. The active-site cavity of HCV NS5B is completely encircled, owing to extensive interactions between the finger and thumb subdomains (Fig. 8). HCV NS5B uses di- or trinucleotides efficiently to initiate RNA replication, whereby the initiation complex, which consists of the polymerase, template and primer, is assembled at the 3′ end of the template RNA77. On the basis of the tertiary structures of HCV NS5B, the RdRp is predicted to be able to accommodate the template–primer duplex without global conformation changes, indicating that the general structure is probably preserved during the reaction pathway. This is in contrast to other previously determined polymerase structures, the inter-subdomain contacts of which are relatively flexible and can undergo large-scale subdomain movement. Recent studies indicate that NS5B can oligomerize, which might be important for modulating the polymerase80. Several groups developed HCV polymerase assays containing recombinant NS5B, for which elongation activity was shown in vitro57,77. In addition to polymerase assays containing only the NS5B protein, RdRp activity can now be analysed in the context of the HCV-replicon system81.
Ribbon representation of the first 570 residues from the amino terminus of hepatitis C virus (HCV) non-structural protein 5B (NS5B), with α-helices and β-strands represented in red and cyan, respectively. The 'thumb', 'finger' and 'palm' subdomains are common to all known polymerases. NH2 and COOH indicate the positions of the amino and carboxy terminus of the protein, respectively. Reproduced, with permission, from Ref. 77 © (2002) Birkhäuser Publishing Ltd.
A series of diketobutanoic acids that have activity against HCV NS5B in vitro and in the HCV-replicon assay have been disclosed82. These compounds apparently interfere with the binding of phosphoryl groups of the nucleotide substrate at the active site of NS5B, thereby preventing the formation of phosphodiester bonds that is catalysed by the polymerase. Inhibition is more potent in the presence of Mn2+ rather than Mg2+ as a divalent cation, which mediates binding and also shows selectivity for the NS5B polymerase. So, diketobutanoic acids might be models for NS5B inhibition in structural studies. In the clinic, the orally active compound JTK-003, which belongs to a class of low-nanomolar, 6,5-fused heterocyclic inhibitors (represented by the benzimidazole derivative compound II in Fig. 9), is now in Phase I and II trials in Japan (Table 2).
This compound represents an analogue of JTK-003, an orally active inhibitor of non-structural protein 5B (NS5B) that is now in Phase I/II trials. Compound II had a half-maximal inhibitory concentration (IC50) of 0.011 mM against hepatitis C virus (HCV) non-structural protein 5B (NS5B) polymerase in vitro. For further details, see Ref. 125.
Recently, a monoclonal antibody that specifically inhibits the RNA-polymerase activity of NS5B was developed83, which might provide insight into the design of other compounds that might inhibit HCV replication. In addition, high-affinity and specific RNA aptamers of NS5B might offer an alternative avenue for inhibiting the RdRp activity84. These results, however, need to be followed up in experimental models before being considered in clinical studies. Finally, some companies are developing nucleoside analogues as inhibitors of HCV NS5B RdRp, driven by the success of nucleoside-analogue inhibitors of HIV reverse transcriptase and the recent proposition that ribavirin, when converted to the triphoshate form, is used by HCV RdRp and causes lethal mutagenesis of the viral genome39.
HCV IRES. The highly conserved regions in the IRES of the HCV RNA genome (Fig. 10), its distinctive translational-initiation mechanism and its essential role in mediating the unusual translational-initiation and replication processes of HCV make these elements an attractive target for compounds that inhibit transcription and translation of the HCV RNA. One drawback to targeting the IRES or the 3′NTR of the HCV genome is that the stem–loop structures, particularly those at the 3′NTR, are very stable, and might, therefore, be difficult to disrupt. However, specific sites for interfering with IRES function have been identified, including subdomains IIb, IIIe and IIId, which interact with the 40S ribosomal subunit and are essential for translation initiation, and subdomain IIIb, which binds the eukaryotic translation-initiation factor eIF3 and is required for IRES-mediated translational activity (Fig. 10). As the tertiary structures of these important subdomains are now available, pharmaceutical companies can apply structure-based methods for the discovery of inhibitors of HCV protein synthesis and replication, as screening efforts for small-molecule inhibitors have not yielded any promising leads.
Shown is the sequence and secondary structure of the 5′ non-translated region (NTR) of the hepatitis C virus (HCV), which contains the internal ribosome-entry site (IRES), including the four domains (I, II, III and IV) and subdomains of domain III (a, b, c, d, e and f). The IRES sequence is indicated in blue, with the location of the start codon (AUG) indicated in red. Numbers refer to nucleotide positions. The binding sites of eukaryotic translation-initiation factor 3 (eIF3) and the 40S ribosomal subunit are indicated in boxes. Solution structures of domain II and subdomains IIIa,b,c, IIId and IIIe are also depicted.
Ribozymes and antisense. Ribozymes have been developed to inhibit HCV replication by cleaving the target HCV genomic RNA. Ribozymes are naturally occurring, short RNA molecules with endoribonuclease activity that can catalyse sequence-specific cleavage of RNA85. The specificity of such catalytic RNA is determined by flanking sequences that are complementary to the target RNA. Heptazyme is such a ribozyme, which targets the HCV IRES and has been shown to inhibit replication of an HCV–poliovirus chimaera in cell culture86. However, Phase II dosing of Heptazyme for the treatment of chronic hepatitis C has been halted owing to blindness that occurred in one animal after receiving high-dose Heptazyme during toxicology testing, although it has not been determined whether the toxicity is due to the drug.
An alternative approach to selectively targeting the HCV RNA genome is the use of antisense oligonucleotide (ASO) technology, which inhibits gene expression by inducing cleavage of the target RNA at the site of oligonucleotide hybridization by an RNaseH-mediated mechanism. Several ASOs that have been designed to bind to the stem–loop structures in the HCV IRES have been effective in inhibiting HCV replication in cell-culture assays and the expression of an HCV luciferase reporter gene in the livers of mice infected with recombinant vaccinia virus expressing the reporter construct87,88. ISIS 14803 is a 20-nucleotide, 5′-methylcytidine phosphorothioate ASO that is in a Phase II clinical trial at present in patients with chronic HCV infections89.
Targeting envelope proteins. The binding of HCV E2 to CD81 and the LDL receptor for viral entry, and the recent development of surrogate models to study virus attachment90,91, also raise the possibility of finding agents to block the binding or entry of the virus into cells. Alternatively, interference with the E2–CD81 interaction might stimulate the INNATE IMMUNE RESPONSE against HCV, as engagement of CD81 by the E2 glycoprotein has been shown recently to inhibit the function of natural-killer cells92,93.
Monoclonal antibodies have the distinctive ability to reduce viral levels by acting directly on the virus, and might be able to prevent re-infection of new liver cells. XTL-002, a fully human, high-affinity monoclonal antibody directed against the HCV E2 protein, is in a Phase Ib clinical study (Table 2). XTL-002 has been shown to reduce, in a dose-dependent manner, serum levels of HCV in the Trimera system16. In Phase Ia clinical trials, more than half of a group of 15 HCV patients who received a single intravenous infusion of XTL-002 experienced significant viral-load reductions, ranging from 2- to 100-fold, without serious side effects.
Vaccines
For developing countries, a preventive vaccine is the only viable option to controlling an emerging viral pandemic. However, vaccine development for HCV has been particularly problematic owing to the presence of large numbers of HCV genotypes and quasispecies. Initial excitement was generated when a chimpanzee that had previously been inoculated with genotype 1a HCV RNA developed immunity to homologous type 1a RNA as well as heterologous 1a strains in re-challenge studies94. Subsequent studies by others showed similar encouraging results95,96, supporting the feasibility of developing a cross-protective vaccine for HCV. Several antigens and delivery strategies, including purified peptide vaccines targeting CTL epitopes and DNA vaccines combined with different adjuvants (for example, CpG dinucleotides and cytokines), as well as recombinant viruses and assembled virus-like particles, are being pursued to stimulate HCV-specific immune responses97,98. Phase IIa clinical studies are underway in Belgium to explore the safety and immune response of an HCV E1 candidate therapeutic vaccine in patients with chronic hepatitis C infection (Table 2).
Emerging and potential new therapies
IMPDH inhibitors. Cellular genes that are required for HCV replication represent an attractive class of genes for small-molecule targeting to control viral replication. On the basis of the success of IFN-α co-therapy with ribavirin, which has been postulated to exert its effects on intracellular pools of guanine nucleotides by inhibiting the cellular IMPDH, new inhibitors of IMPDH are being developed for the potential treatment of HCV infection. VX-497 (merimepodib) (Fig. 11) is the lead compound in a series of oral IMPDH inhibitors and is in Phase II clinical development99. VX-497 has the potential to exert direct antiviral activity, and it has been shown to effect lymphocyte migration and proliferation involved in the immune-system response, indicating that it has the potential to treat both viral proliferation and liver inflammation.
VX-497 is an oral inosine monophosphate dehydrogenase (IMPDH) inhibitor that is in Phase II clinical trials for hepatitis C virus (HCV) treatment. A Phase II trial of the triple combination therapy of VX-497, PEGylated interferon-α (PEG-IFN-α) and ribavirin is underway (Table 1).
Ribavirin analogues. Two new analogues of ribavirin are under development (Table 2). Levovirin (Fig. 3) is the l-enantiomer of ribavirin, and shares similar immunomodulatory activities with ribavirin, but has a more favourable in vivo toxicology and better tolerability profiles. However, Levovirin has no in vitro antiviral activity against a panel of RNA and DNA viruses, indicating that structural modification of ribavirin can dissociate its immunomodulatory properties from its antiviral and toxicological properties. Viramidine is a liver-targeting analogue of ribavirin; it is an inactive PRODRUG of ribavirin that is activated by deamination in the liver by adenosine deaminase (Fig. 3). Initial results from a Phase I trial indicated that the prodrug had slightly less haematological toxicity than ribavirin and was well tolerated. In addition, Viramidine was more efficiently targeted to the liver in rats, with a remarkably decreased concentration in red blood cells compared with ribavirin.
Glucosidase inhibitors. During viral assembly, mammalian viruses are dependent on the host-cell glycosylation machinery for morphogenesis and secretion100. The cellular enzymes that mediate the first step in the N-linked glycosylation pathway are the ER-localized glucosidases, including ER α-glucosidases I and II. Imino-sugar derivatives, such as N-butyldeoxynojirimycin (NB-DNJ) and N-nonyl-deoxynojirimycin (NN-DNJ), which competitively inhibit ER glucosidases, have been shown to block replication of woodchuck hepatitis virus (WHV)101,102 and bovine diarrhea virus (BVDV), an in vitro surrogate model of hepatitis C103,104,105, as well as the flaviviruses dengue-virus serotype 2 and Japanese-encephalitis virus106. So, targeting ER α-glucosidases at a low level could be a potential strategy for treating viral infections without compromising the host cell. The potential use of imino sugars as broad-spectrum anti-hepatitis-virus agents has been reviewed recently107.
Inhibition of replicase/ER association. The NS proteins of HCV are believed to form a membrane-associated replicase together with unidentified host components, as shown by their association with the ER membrane108. Association of NS5B with the ER membrane is mediated by the carboxyl-terminal 21 amino-acid residues that are highly conserved among HCV isolates, and are predicted to form a transmembrane α-helix109. The membrane-anchor region of the NS5A phosphoprotein has been mapped to the amino-terminal 30 amino-acid residues, which contain an amphipathic α-helix that is also highly preserved among HCV isolates110. The hydrophobic amino-terminal domain of NS4A is required for ER targeting of NS3 (Ref. 111). So, formation of the HCV replicase seems to involve specific determinants for membrane association, which might represent novel targets for antiviral intervention.
Inhibition of viral assembly/transport. Topology and subcellular-localization studies indicate that the p7 polypeptide might have a functional role in several compartments of the secretory pathway, possibly in the production of progeny virus or virion assembly22. In fact, mutation of the equivalent protein of BVDV can inhibit the production of infectious virus112. Recent studies have shown that p7 can form hexamers, particularly in the presence of lipid membranes, enhancing membrane permeability113. The hexameric forms of p7 resemble cation-channel pores, which are reminiscent of viroporins, a family of short, viral integral polypeptides that can form a hydrophilic pore in the membrane by oligomerization and subsequently cause membrane destabilization112. Intriguingly, amantadine, which was evaluated in combination with IFN-α and ribavirin for chronic hepatitis C treatment45, can reverse the membrane permeability effects of HCV p7 (Ref. 112). If these results can be substantiated in an experimental model, HCV p7 could be a potential target for therapeutic intervention for blocking the production of infectious HCV progeny.
RNA-mediated interference. RNA interference (RNAi) is an emerging powerful technology for sequence-specific targeting and degradation of messenger RNA114,115. Early studies in plants initially established RNAi or RNA silencing as part of a natural antiviral defence, a mechanism that has now been extended to animals. The proposed mechanism involves cleavage of dsRNA, introduced as either a viral replicative intermediate or an artificial construct, into small interfering RNA (siRNA) fragments of 21–22 base pairs that target mRNA of homologous sequence for specific degradation. Importantly, several recent reports have shown specific inhibitory effects of synthetic siRNA on the replication of HIV-1 (Refs 116–118) and poliovirus119 in cultured cells. siRNA-induced inhibition of the HCV NS5B gene was recently shown using co-transfection studies in adult mice120. However, the feasibility and efficacy of siRNA remain to be tested in animals infected with live virus. A major limitation of the use of siRNA as an antiviral therapeutic is that the effects are transient, as mammals, in contrast to worms and plants, apparently lack the mechanisms that amplify the silencing cycles. So, the development of an efficient delivery system will be crucial for the use of RNAi as a therapeutic. Despite these challenges, the rapid progress of the RNAi technology has opened up new avenues for novel anti-HCV therapies.
Liver damage and hepatocellular carcinoma. In addition to drugs with direct antiviral properties, therapies that preserve the cell structure or prevent/reverse the fibrosis and cirrhosis caused by chronic hepatitis C are also needed. Recent data indicate that fibrosis, and perhaps even early cirrhosis, might be reversible to some extent. Furthermore, improved liver histology might enhance the subsequent response to IFN-based therapies. In a small pilot study, interleukin-10 (IL-10), a cytokine that downregulates the pro-inflammatory response and modulates hepatic fibrogenesis, was found to reduce the degree of liver fibrosis and improve liver histology in patients with chronic hepatitis C who were refractory to treatments with IFN alone or in combination with ribavirin121 (Table 2). IP-501 is an orally administered antifibrotic compound that is being tested for the treatment of alcohol-induced liver disease and chronic hepatitis-C-induced cirrhosis, whereas Actimmune, an anti-IFN-γ, is now undergoing a Phase II trial for the treatment of severe liver fibrosis or cirrhosis caused by HCV (Table 2). Excessive apoptosis (programmed cell death) of the liver is another hallmark characteristic of hepatitis C, so drugs that modulate the apoptotic pathway could be used to treat the disease122. To this end, a caspase inhibitor (IDN-6556), which has a hepatoprotective effect for hepatitis C treatment, is in Phase II trials (Table 2). Drugs are also in need to treat hepatocellular carcinoma, which is on the rise in the United States primarily owing to increased incidence of HCV infection rates. Clinical results indicate that the compound T67, which binds irreversibly to β-tubulin, might have particular relevance for the treatment of this aggressive form of cancer.
Recurrent hepatitis C after liver-transplant surgery. Better agents are also needed to prevent the recurrence of hepatitis C after liver-transplant surgery, which is a significant unmet medical need5. Re-infection by HCV is nearly universal in patients who have received liver transplants due to chronic HCV infection. Antiviral therapy has been used to prevent or treat recurrent hepatitis C, but tolerance is poor in post-transplant patients. The immunosupressive drug CellCept (mycophenolate mofetil; Fig. 12), when used at high doses for at least one year after liver transplantation, was found to reduce the incidence of hepatitis C re-infection (Table 2). Interestingly, mycophenolate mofetil inhibits IMPDH but seems to a have a weak effect on HCV replication, and studies are planned for its use in combination with PEGylated IFN-α to treat HCV infection. A Phase I/II clinical trial has been initiated to evaluate Civacir or hepatitis C immunoglobulin for preventing HCV recurrence of transplanted livers in patients suffering from hepatitis C (Table 2).
Summary and concluding remarks
The clinical pipeline for hepatitis C is showing promise for safer and more effective therapies; most of these focus on modification of the IFN molecule (Tables 1,2). Inhibitors in Phase II clinical development include those that target the HCV IRES, NS3 protease and NS5B polymerase. Some companies are focusing their efforts on developing ribavirin analogues, whereas others are exploring host drug discovery targets. For those pathways that are vital to fundamental cellular processes, modulation rather than ablation of the enzymes that are involved is likely to be a necessity. One of the biggest challenges remains the development of an effective and safe prophylactic vaccine, and failing that, a therapeutic vaccine. We will need to understand better how HCV interacts with and evades the adaptive immune responses, which might aid in the development of universally effective therapies and vaccines for HCV. We know it is possible to stimulate the immune system to attack HCV, as shown by IFN-α, one of the best known of such immunomodulators. Bearing in mind that the use of immune modulators might have undesired effects in HCV/HIV co-infected patients, which will need to be addressed carefully with proper research before clinical studies, it is hoped that other immune modulators will be developed as we analyse the ever-complex interplay between HCV and the host immune system and the mechanisms of HCV resistance to IFN. In addition, we need to understand better the mechanisms by which HCV induces liver cirrhosis, fibrosis and hepatocellular carcinoma, as well as its role in extrahepatic infections, to develop a more comprehensive treatment of hepatitis C.
References
Choo, Q. L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989). The original report to describe the molecular cloning of the HCV virus from a complementary DNA library made from blood containing the non-A, non-B hepatitis agent.
Hoofnagle, J. H. Hepatitis C — the clinical spectrum of disease. Hepatology 26, S15–S20 (1997).
Memon, M. I. & Memon, M. A. Hepatitis C: an epidemiological review. J. Viral Hepatitis 9, 84–100 (2002).
Anonymous. Hepatitis C — global prevalence (update). World Health Org. Weekly Epidemiol. Rec. 75, 18–19 (2000).
Willems, M., Metselaar, H. J., Tilanus, H. W., Schalm, S. W. & de Man, R. A. Liver transplantation and hepatitis C. Transplant Int. 15, 61–72 (2002).
Pollard, R. B. Analogy of human immunodeficiency virus to hepatitis C virus: the human immunodeficiency model. Am. J. Med. 107, 41S–44S (1999).
Rehermann, B. Interaction between the hepatitis C virus and the immune system. Semin. Liv. Dis. 20, 127–141 (2000).
Di Bisceglie, A. M., McHutchinson, J. & Rice, C. M. New therapeutic strategies for hepatitis C. Hepatology 35, 224–231 (2002). An excellent short review about the most recent developments in HCV therapeutic strategies.
Wang, Q. M. & Heinz, B. A. Recent advances in prevention and treatment of hepatitis C virus infections. Prog. Drug Res. 55, 1–32 (2000).
Dymock, B. W., Jones, P. S. & Wilson, F. X. Novel approaches to the treatment of hepatitis C virus infection. Antivir. Chem. Chemother. 11, 79–96 (2000).
Ideo, G. & Bellobuono, A. New therapies for the treatment of chronic hepatitis C. Curr. Pharm. Des. 8, 959–966 (2002).
Locarnini, S. A. & Bartholomeusz, A. Advances in hepatitis C: what is coming in the next 5 years? J. Gastroenterol. Hepatol. 17, 442–447 (2002).
Myles, D. C. Recent advances in the discovery of small molecule therapies for HCV. Curr. Opin. Drug Discov. Dev. 4, 411–416 (2001). A review about the advances in small-molecule antivirals for the treatment of HCV.
Bartenschlager, R. & Lohmann, V. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 52, 1–17 (2001). A comprehensive review about the developments in HCV cell-culture systems, including the replicon cell-based system, written by the inventors of the HCV-replicon system.
Mercer, D. F. et al. Hepatitis C virus replication in mice with chimeric human livers. Nature Med. 7, 927–933 (2001). A description of an elegant and novel mouse model that supports prolonged HCV infection, which could be serially passaged through three generations of mice, confirming both synthesis and release of infectious viral particles. This is the first mouse model to be suitable for studying human HCV in vivo.
Ilan, E. et al. The hepatitis C virus (HCV)-Trimera mouse: a model for evaluation of agents against HCV. J. Infect. Dis. 185, 153–161 (2002).
Robertson, B. et al. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol. 143, 2493–2503 (1998).
Rosenberg, S. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313, 451–464 (2001).
Friebe, P., Lohmann, V., Krieger, N. & Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75, 12047–12057 (2001). A description of sequences that are necessary for HCV RNA replication, analysing functions of the different 3′NTR domains.
Friebe, P. & Bartenschlager, R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76, 5326–5338 (2002).
Reed, K. E. & Rice, C. M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Hep. C Viruses 242, 55–84 (2000).
Carrere-Kremer, S. et al. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 76, 3720–3730 (2002).
Walewski, J. L., Keller, T. R., Stump, D. D. & Branch, A. D. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA-A Pub. RNA Soc. 7, 710–721 (2001).
Xu, Z. M. et al. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20, 3840–3848 (2001).
Shirota, Y. et al. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277, 11149–11155 (2002).
Bartenschlager, R. & Lohmann, V. Replication of hepatitis C virus. J. Gen. Virol. 81, 1631–1648 (2000). A review about all aspects of the HCV life cycle and model systems.
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998).
Monazahian, M. et al. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57, 223–229 (1999).
Agnello, V., Abel, G., Elfahal, M., Knight, G. B. & Zhang, Q. X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl Acad. Sci. USA 96, 12766–12771 (1999).
Neumann, A. U. et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-α therapy. Science 282, 103–107 (1998). A solid study of HCV dynamics in patients before and after IFN treatment.
Bukh, J., Miller, R. H. & Purcell, R. H. Genetic heterogeneity of hepatitis C virus — quasispecies and genotypes. Semin. Liv. Dis. 15, 41–63 (1995).
Zein, N. N. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13, 223–235 (2000).
Herion, D. & Hoofnagle, J. H. The interferon sensitivity determining region — all hepatitis C virus isolates are not the same. Hepatology 25, 769–771 (1997).
Tan, S. L. & Katze, M. G. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284, 1–12 (2001).
Farci, P. & Purcell, R. H. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liv. Dis. 20, 103–126 (2000). A review that describes the effects of HCV genotypes and quasispecies on treatment outcome in the clinic.
Dammacco, F. et al. The lymphoid system in hepatitis C virus infection: autoimmunity, mixed cryoglobulinemia, and overt B-cell malignancy. Semin. Liv. Dis. 20, 143–157 (2000).
Hoofnagle, J. H. et al. Treatment of chronic non-A, non-B hepatitis with recombinant human α-interferon. A preliminary report. N. Engl. J. Med. 315, 1575–1578 (1986).
Scott, L. J. & Perry, C. M. Interferon-α2b plus ribavirin — a review of its use in the management of chronic hepatitis C. Drugs 62, 507–556 (2002).
Lau, J. Y. N., Tam, R. C., Liang, T. J. & Hong, Z. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 35, 1002–1009 (2002).
Bekkering, F. C., Brouwer, J. T., Hansen, B. E. & Schalm, S. W. Hepatitis C viral kinetics in difficult to treat patients receiving high dose interferon and ribavirin. J. Hepatol. 34, 435–440 (2001).
Tsutsumi, M., Takada, A., Takase, S. & Sawada, M. Effects of combination therapy with interferon and ofloxacin on chronic type C hepatitis — a pilot study. J. Gastroenterol. Hepatol. 11, 1006–1011 (1996).
Komatsu, M. et al. Pilot study of ofloxacin and interferon-α combination therapy for chronic hepatitis C without sustained response to initial interferon administration. Can. J. Gastroenterol. 11, 507–511 (1997).
Moscarella, S. et al. Interferon and thymosin combination therapy in naive patients with chronic hepatitis C — preliminary results. Liver 18, 366–369 (1998).
Brillanti, S., Levantesi, F., Masi, L., Foli, M. & Bolondi, L. Triple antiviral therapy as a new option for patients with interferon nonresponsive chronic hepatitis C. Hepatology 32, 630–634 (2000).
Zilly, M. et al. Triple antiviral re-therapy for chronic hepatitis C with interferon-α, ribavirin and amantadine in nonresponders to interferon-α and ribavirin. Eur. J. Med. Res. 7, 149–154 (2002).
Kozlowski, A., Charles, S. A. & Harris, J. M. Development of pegylated interferons for the treatment of chronic hepatitis C. Biodrugs 15 419–429 (2001).
Zeuzem, S. et al. PEGinterferon-α2a in patients with chronic hepatitis C. N. Engl. J. Med. 343, 1666–1672 (2000).
Heathcote, E. J. et al. PEG-interferon-α2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343, 1673–1680 (2000).
Manns, M. P. et al. PEG-interferon-α2b plus ribavirin compared with interferon-α2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358, 958–965 (2001). References 47–49 describe results of a clinical trial that was done with PEGylated IFN alone or in combination with ribavirin, compared with unmodified IFN.
Enomoto, M. et al. Dynamics of hepatitis C virus monitored by real-time quantitative polymerase chain reaction during first 2 weeks of IFN-β treatment are predictive of long-term therapeutic response. J. Interferon Cytokine Res. 22, 389–395 (2002).
Barbaro, G. et al. Intravenous recombinant interferon-β versus interferon-α2b and ribavirin in combination for short-term treatment of chronic hepatitis C patients not responding to interferon-α. Scand. J. Gastroenterol. 34, 928–933 (1999).
Kumashiro, R. et al. Interferon-γ brings additive anti-viral environment when combined with interferon-α in patients with chronic hepatitis C. Hepatol. Res. 22, 20–26 (2002).
Barbaro, G. & Barbarini, G. Consensus interferon for chronic hepatitis C patients with genotype 1 who failed to respond to, or relapsed after, interferon-α2b and ribavirin in combination: an Italian pilot study. Eur. J. Gastroenterol. Hepatol. 14, 477–483 (2002).
Chang, C. C. et al. Evolution of a cytokine using DNA family shuffling. Nature Biotechnol. 17, 793–797 (1999).
Maier, I. & Wu, G. Y. Hepatitis C and HIV co-infection: a review. World J. Gastroenterol. 8, 577–579 (2002).
Falck-Ytter, Y. et al. Surprisingly small effect of antiviral treatment in patients with hepatitis C. Ann. Intern. Med. 136, 288–292 (2002).
De Francesco, R. et al. Biochemical and immunologic properties of the nonstructural proteins of the hepatitis C virus: implications for development of antiviral agents and vaccines. Sem. Liv. Dis. 20, 69–83 (2000). A review that describes the function and properties of HCV NS proteins and their implications for drug and vaccine discovery.
Darke, P. L., Jacobs, A. R., Waxman, L. & Kuo, L. C. Inhibition of hepatitis C virus NS2/3 processing by NS4A peptides — implications for control of viral processing. J. Biol. Chem. 274, 34511–34514 (1999).
Thibeault, D., Maurice, R., Pilote, L., Lamarre, D. & Pause, A. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276, 46678–46684 (2001).
Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M. & Rice, C. M. A second hepatitis C virus-encoded proteinase. Proc. Natl Acad. Sci. USA 90, 10583–10587 (1993).
Hijikata, M. et al. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67, 4665–4675 (1993).
Whitney, M. et al. A collaborative screening program for the discovery of inhibitors of HCV NS2/3 cis-cleaving protease activity. J. Biomol. Screening 7, 149–154 (2002). References 58–62 describe the identification, in vivo validation and in vitro assay development for the NS2–NS3 protease activity of HCV. It is now possible to screen this novel protease target for small-molecule inhibitors.
Kolykhalov, A. A., Mihalik, K., Feinstone, S. M. & Rice, C. M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74, 2046–2051 (2000).
Yao, N. H., Reichert, P., Taremi, S. S., Prosise, W. W. & Weber, P. C. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7, 1353–1363 (1999).
Ingallinella, P. et al. Prime site binding inhibitors of a serine protease: NS3/4A of hepatitis C virus. Biochemistry 41, 5483–5492 (2002).
Zhang, R. M., Durkin, J. P. & Windsor, W. T. Azapeptides as inhibitors of the hepatitis C virus NS3 serine protease. Bioorg. Med. Chem. Lett. 12, 1005–1008 (2002).
Slater, M. J. Design and synthesis of pyrrolidine-5,5-trans-lactams as novel inhibitors of hepatitis C virus protease. Antiviral Research — 15th International Conference, Prague, Czech Rebublic (2002).
Fukuda, K. et al. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 267, 3685–3694 (2000).
Kumar, P. K. R. et al. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology 237, 270–282 (1997).
Yao, N. H. et al. Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol. 4, 463–467 (1997).
Kang, L. W. et al. Crystallization and preliminary X-ray crystallographic analysis of the helicase domain of hepatitis C virus NS3 protein. Acta Crystallograph. D 54, 121–123 (1998).
Levin, M. K. & Patel, S. S. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274, 31839–31846 (1999).
Pang, P. S., Jankowsky, E., Planet, P. J. & Pyle, A. M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21, 1168–1176 (2002).
Tanji, Y., Hijikata, M., Satoh, S., Kaneko, T. & Shimotohno, K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69, 1575–1581 (1995).
Yang, S. H., Lee, C. G., Song, M. K. & Sung, Y. C. Internal cleavage of hepatitis C virus NS3 protein is dependent on the activity of NS34A protease. Virology 268, 132–140 (2000).
Kolykhalov, A. A. et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277, 570–574 (1997). A description of an infectious HCV clone that can transmit HCV by injection of its RNA into the liver of chimpanzees. This clone was infectious only after a consensus clone was derived from many patient isolates, implying that not every virus in a patient is infectious. This work defines the structure of a functional HCV-genome RNA, and proves that HCV alone is sufficient to cause disease.
Leveque, V. J. -P. & Wang, Q. M. RNA-dependent RNA polymerase encoded by hepatitis C virus: biomedical applications. Cell. Mol. Life Sci. 59, 909–919 (2002).
Bressanelli, S. et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA 96, 13034–13039 (1999).
Lesburg, C. A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol. 6, 937–943 (1999).
Qin, W. P. et al. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277, 2132–2137 (2002).
Cheney, I. W. et al. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297, 298–306 (2002).
De Francesco, R. Diketobutanoic acids as HCV Pol inhibitors. 7th International Meeting on Hepatitis C Virus and Related Viruses (Molecular Virology and Pathogenesis), Gold Coast, Queensland, Australia (2000).
Moradpour, D. et al. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277, 593–601 (2002). A description of a monoclonal antibody that potently and specifically inhibits HCV RdRP in vitro . This could be a new avenue towards developing an HCV therapeutic.
Biroccio, A., Hamm, J., Incitti, I., De Francesco, R. & Tomei, L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76, 3688–3696 (2002).
Usman, N. & Blatt, L. R. M. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J. Clin. Invest. 106, 1197–1202 (2000).
Macejak, D. G. et al. Inhibition of hepatitis C virus (HCV)-RNA-dependent translation and replication of a chimeric HCV poliovirus using synthetic stabilized ribozymes. Hepatology 31, 769–776 (2000).
Zhang, H. et al. Antisense oligonucleotide inhibition of hepatitis C virus (HCV) gene expression in livers of mice infected with an HCV vaccinia virus recombinant. Antimicrob. Agents Chemother. 43, 347–353 (1999).
Brown-Driver, V., Eto, T., Lesnik, E., Anderson, K. P. & Hanecak, R. C. Inhibition of translation of hepatitis C virus RNA by 2′-modified antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 9, 145–154 (1999).
Witherell, G. ISIS-14803 (Isis Pharmaceuticals). Curr. Opin. Invest. Drugs 2, 1523–1529 (2001).
Lambot, M. et al. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277, 20625–20630 (2002).
Wellnitz, S. et al. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76, 1181–1193 (2002).
Tseng, C. T. K. & Klimpel, G. R. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195, 43–49 (2002).
Crotta, S. et al. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195, 35–41 (2002).
Weiner, A. J. et al. Intrahepatic genetic inoculation of hepatitis C virus RNA confers cross-protective immunity. J. Virol. 75, 7142–7148 (2001).
Major, M. E. et al. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J. Virol. 76, 6586–6595 (2002).
Bassett, S. E. et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology 33, 1479–1487 (2001).
Bukh, J., Forns, X., Emerson, S. U. & Purcell, R. H. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology 44, 132–142 (2001).
Lechmann, M. & Liang, T. J. Vaccine development for hepatitis C. Semin. Liv. Dis. 20, 211–226 (2000).
Markland, W., McQuaid, T. J., Jain, J. & Kwong, A. D. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with α-interferon. Antimicrob. Agents Chemother. 44, 859–866 (2000).
Dwek, R. A., Butters, T. D., Platt, F. M. & Zitzmann, N. Targeting glycosylation as a therapeutic approach. Nature Rev. Drug Discov. 1, 65–75 (2002).
Block, T. M. et al. Treatment of chronic hepadnavirus infection in a woodchuck animal model with an inhibitor of protein folding and trafficking. Nature Med. 4, 610–614 (1998).
Mehta, A. et al. Inhibition of hepatitis B virus DNA replication by imino sugars without the inhibition of the DNA polymerase: therapeutic implications. Hepatology 33, 1488–1495 (2001).
Zitzmann, N. et al. Imino sugars inhibit the formation and secretion of bovine viral diarrhea virus, a pestivirus model of hepatitis C virus: implications for the development of broad spectrum anti-hepatitis virus agents. Proc. Natl Acad. Sci. USA 96, 11878–11882 (1999).
Durantel, D. et al. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 75, 8987–8998 (2001).
Jordan, R. et al. Inhibition of host ER glucosidase activity prevents Golgi processing of virion-associated bovine viral diarrhea virus E2 glycoproteins and reduces infectivity of secreted Virions. Virology 295, 10–19 (2002).
Wu, S. F. et al. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 76, 3596–3604 (2002).
Block, T. M. & Jordan, R. Iminosugars as possible broad spectrum anti hepatitis virus agents: the glucovirs and alkovirs. Antiviral Chem. Chemother. 12, 317–325 (2001).
Mottola, G. et al. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293, 31–43 (2002).
Schmidt-Mende, J. et al. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276, 44052–44063 (2001).
Brass, V. et al. An amino-terminal amphipathic α-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277, 8130–8139 (2002).
Wolk, B. et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74, 2293–2304 (2000).
Harada, T., Tautz, N. & Thiel, H. J. E2–p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J. Virol. 74, 9498–9506 (2000).
Griffin, S. Hexamerization of the hepatitis C virus p7 protein: possible fuction as a virus-encoded cation channel. 9th International Meeting on Hepatitis and Related Viruses, San Diego, USA (2002).
Hannon, G. J. RNA interference. Nature 418, 244–251 (2002).
Zamore, P. D. Ancient pathways programmed by small RNAs. Science 296, 1265–1269 (2002).
Novina, C. D. et al. siRNA-directed inhibition of HIV-1 infection. Nature Med. 8, 681–686 (2002).
Lee, N. S. et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nature Biotechnol. 20, 500–505 (2002).
Jacque, J. M., Triques, K. & Stevenson, M. Modulation of HIV-1 replication by RNA interference. Nature 418, 435–438 (2002).
Gitlin, L., Karelsky, S. & Andino, R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418, 430–434 (2002).
McCaffrey, A. P. et al. Gene expression — RNA interference in adult mice. Nature 418, 38–39 (2002).
Nelson, D. R., Lauwers, G. Y., Lau, J. Y. N. & Davis, G. L. Interleukin 10 treatment reduces fibrosis in patients with chronic hepatitis C: a pilot trial of interferon nonresponders. Gastroenterology 118, 655–660 (2000).
Muratori, L. & Gibellini, D. A new route to apoptosis in hepatitis C virus infection. J. Hepatol. 35, 814–815 (2001).
Tsantrizos, Y. S. et al. Macrocyclic peptides active against the hepatitis C virus. Int. Patent Appl. WO 00/59929 (2000).
Kim, J. L. et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 (1998).
Hashimoto, H. et al. Preparation of heterocyclic compounds as remedies for hepatitis C. Int. Patent Appl. WO 01/47883 (2001).
Acknowledgements
We would like to thank J. Colacino, M. López Lastra and our anonymous reviewers for reading this manuscript and providing helpful suggestions, and K. McKnight, M. Bures, J. Puglisi, Y. S. Tsantrizos, J. Tang, P. l. Caron, M. Wang and K.-L. Yu for providing the various Figures. Unfortunately, owing to space constraints, many citations and explanations have been limited.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Cancer.gov
LocusLink
FURTHER INFORMATION
Encyclopedia of Life Sciences
Glossary
- FIBROSIS
-
A process that replaces lost parenchymal tissue, resulting in scar formation.
- CIRRHOSIS
-
A liver disease in which parenchymal tissues die and the liver becomes filled with fibrous tissue.
- HEPATOCELLULAR CARCINOMA
-
A malignant tumour of the liver that is seen in some people with long-term liver damage due to chronic hepatitis C or hepatitis B.
- VIRAL LOAD
-
The amount of virus that is present in the blood.
- CYTOTOXIC T LYMPHOCYTES
-
A subset of T lymphocytes that can kill body cells that have been infected by viruses or transformed by cancer.
- TH1 CELLS
-
T lymphocytes that produce cytokines to help inflammation and antiviral responses.
- TH2 CELLS
-
T lymphocytes that produce cytokines to help antibody responses.
- INTERFERONS
-
(IFN). Secreted cytokines that are known for their antiviral, antiproliferative and immunomodulatory activities. There are two types of IFN: type I (IFN-α, IFN-β, IFN-ω and IFN-τ) and type II (IFN-γ). There are at least 14 IFN-α genes, but only one IFN-β and IFN-γ gene has been reported so far.
- HCV REPLICON
-
A biscistronic DNA construct that contains a selectable marker gene and genes encoding HCV non-structural proteins, in which an HCV IRES and an EMCV IRES direct the translation of the marker gene and viral genes, respectively. Transfection of RNA transcribed from such constructs into the hepatoma cell line Huh-7 results in selectable, autonomously replicating HCV RNAs.
- ENVELOPE
-
A lipoprotein-bilayer outer membrane of many viruses. Often heavily glycosylated, envelope proteins usually function to identify and attach the virus to the cell-surface receptor, so that viral entry can occur.
- NUCLEOCAPSID
-
The coat (capsid) of a virus plus the enclosed nucleic-acid genome.
- VIRION
-
A mature infectious virus particle that exists freely outside the host cell.
- NUCLEOSIDE TRIPHOSPHATASE
-
An enzyme that hydrolyses nucleoside triphosphate (NTP) — a nucleotide that is of fundamental importance as a carrier of chemical energy in all living organisms — to drive energetically unfavourable biological processes.
- RNA HELICASE
-
An ATP-dependent enzyme that catalyses the unwinding of RNA helices.
- ENDOCYTOSIS
-
A process by which proteins or viral particles at the cell surface are internalized, being transported into the cell within membranous vesicles.
- NEGATIVE-STRAND RNA
-
Genomic viral RNA that is complementary to the messenger RNA (positive strand) that is produced during replication.
- PKR
-
A serine/threonine protein kinase and among the best-studied effectors of the host interferon (IFN)-induced antiviral and antiproliferative response system. In response to stress signals, including virus infection, the normally latent PKR becomes activated through autophosphorylation and dimerization and phosphorylates the eIF2α translation-initiation-factor subunit, leading to an inhibition of the intiation of mRNA translation.
- QUASISPECIES
-
A family of closely related, but slightly different, viral genomes. Viral genetic variants, derived from the original infecting virus, which are present during an infection.
- MIXED CRYOGLOBULINAEMIA
-
The presence of abnormal proteins called cryoglobulins in blood. Cryoglobulinaemia can cause damage to the kidneys.
- GLOMERULONEPHRITIS
-
A kidney disease that affects the capillaries of the glomeruli (the compact cluster of capillaries in the kidney that filter blood) — characterized by oedema, raised blood pressure and excess protein in the urine.
- LEUKOPAENIA
-
A decrease in the number of leukocytes (white blood cells).
- THROMBOCYTOPAENIA
-
A condition in which there is an abnormally small number of platelets in the circulating blood.
- SUSTAINED VIROLOGICAL RESPONSE
-
(SVR). The continued lack of detectable serum HCV RNA six months after the completion of treatment.
- LETHAL MUTAGENESIS
-
A process by which animal RNA viruses, given their high mutation frequencies, undergo a sharp decline in viability after a modest increase in mutation frequency that results from the promiscuous incorporation of nucleoside analogues, such as ribavirin triphosphate, by the viral RNA polymerase.
- HAEMOLYTIC ANAEMIA
-
A decrease in the normal level of erythocytes (red blood cells) in the bloodstream owing to the destruction (rather than underproduction) of red blood cells.
- PEGYLATION
-
A technique of conjugating polyethylene glycol (PEG) groups to proteins to increase their resistance to proteolytic degradation, improve their water solubility and reduce their antigenicity.
- ALBUMIN
-
A protein that is made in the liver, and the most abundant protein in the blood. A low albumin level is associated with liver cirrhosis.
- MOLECULAR BREEDING
-
In vitro recombination-based directed evolution to yield a high percentage of functional variants, as identified and evaluated using marker-assisted selection.
- PEPTIDOMIMETIC
-
A compound containing non-peptidic structural elements that can mimic or antagonize the biological action(s) of a natural parent peptide.
- STRUCTURE–ACTIVITY RELATIONSHIP
-
(SAR). The relationship between chemical structure and pharmacological activity for a series of compounds. Compounds are often classed together because they have structural characteristics in common, including shape, size, stereochemical arrangement and distribution of functional groups. Other factors contributing to the structure–activity relationship include chemical reactivity, electronic effects, resonance and inductive effects.
- RNA APTAMER
-
A single-stranded RNA oligonucleotide that assumes a specific, sequence-dependent shape and binds to a target protein on the basis of a lock-and-key fit between the two molecules.
- INNATE IMMUNE RESPONSE
-
A crucial response during the early phase of host defence against infection by pathogens, before the antigen-specific, adaptive immune response is induced.
- PRODRUG
-
A pharmacologically inactive compound that is converted to the active form of the drug by endogenous enzymes or metabolism. It is generally designed to overcome problems that are associated with stability, toxicity, lack of specificity or limited (oral) bioavailability.
Rights and permissions
About this article
Cite this article
Tan, SL., Pause, A., Shi, Y. et al. Hepatitis C therapeutics: current status and emerging strategies. Nat Rev Drug Discov 1, 867–881 (2002). https://doi.org/10.1038/nrd937
Issue Date:
DOI: https://doi.org/10.1038/nrd937
This article is cited by
-
A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites
Nature Communications (2019)
-
Chemical genetics-based development of small molecules targeting hepatitis C virus
Archives of Pharmacal Research (2017)
-
Identification of novel inhibitors of HCV NS3 protease genotype 3 subtype B through molecular docking studies of phytochemicals from Boerhavia diffusa L
Medicinal Chemistry Research (2017)
-
Structure–activity relationship studies on quinoxalin-2(1H)-one derivatives containing thiazol-2-amine against hepatitis C virus leading to the discovery of BH6870
Molecular Diversity (2015)
-
k nearest neighbor-molecular field analysis on human HCV NS5B polymerase inhibitors: 2,5-disubstituted imidazo[4,5-c]pyridines
Medicinal Chemistry Research (2013)