Abstract
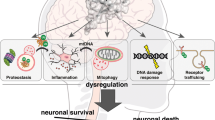
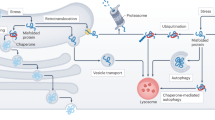
Neurodegenerative disorders of ageing (NDAs) such as Alzheimer disease, Parkinson disease, frontotemporal dementia, Huntington disease and amyotrophic lateral sclerosis represent a major socio-economic challenge in view of their high prevalence yet poor treatment. They are often called 'proteinopathies' owing to the presence of misfolded and aggregated proteins that lose their physiological roles and acquire neurotoxic properties. One reason underlying the accumulation and spread of oligomeric forms of neurotoxic proteins is insufficient clearance by the autophagic–lysosomal network. Several other clearance pathways are also compromised in NDAs: chaperone-mediated autophagy, the ubiquitin–proteasome system, extracellular clearance by proteases and extrusion into the circulation via the blood–brain barrier and glymphatic system. This article focuses on emerging mechanisms for promoting the clearance of neurotoxic proteins, a strategy that may curtail the onset and slow the progression of NDAs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Menzies, F. M. et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017).
Ciechanover, A. & Kwon, Y. T. Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 11, 185 (2017).
Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017).
Galluzzi, L. et al. Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 (2017).
Galluzzi, L., Bravo- San Pedro, J. M., Levine, B., Green, D. R. & Kroemer, G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487–511 (2017).
Ferrer, I. Diversity of astroglial responses across human neurodegenerative disorders and brain aging. Brain Pathol. 27, 645–674 (2017).
Yeh, F. L., Hansen, D. V. & Sheng, M. TREM2, microglia, and neurodegenerative diseases. Trends Mol. Med. 23, 512–533 (2017).
Jansen, A. H., Reits, E. A. & Hol, E. M. The ubiquitin proteasome system in glia and its role in neurodegenerative diseases. Front. Mol. Neurosci. 7, 73 (2014).
Kerr, J. S. et al. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 40, 151–166 (2017).
Molino, D., Zemirli, N., Codogno, P. & Morel, E. The journey of the autophagosome through mammalian cell organelles and membranes. J. Mol. Biol. 429, 497–514 (2017).
Wei, Y. et al. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238.e10 (2017).
Khaminets, A., Behl, C. & Dikic, I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 (2016).
Banerjee, K., Munshi, S., Frank, D. E. & Gibson, G. E. Abnormal glucose metabolism in alzheimer's disease: relation to autophagy/mitophagy and therapeutic approaches. Neurochem. Res. 40, 2557–2569 (2015).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Fekadu, J. & Rami, A. Beclin-1 deficiency alters autophagosome formation, lysosome biogenesis and enhances neuronal vulnerability of HT22 hippocampal cells. Mol. Neurobiol. 53, 5500–5509 (2016).
Lee, S., Sato, Y. & Nixon, R. A. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J. Neurosci. 31, 7817–7830 (2011).
Boland, B. et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 28, 6926–6937 (2008).
Nixon, R. A. The role of autophagy in neurodegenerative disease. Nat. Med. 19, 983–997 (2013).
Rubinsztein, D. C., Codogno, P. & Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 (2012).
Fullgrabe, J., Klionsky, D. J. & Joseph, B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 15, 65–74 (2014).
Herzig, S. & Shaw, R. J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135 (2017).
Fullgrabe, J., Ghislat, G., Cho, D. H. & Rubinsztein, D. C. Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 129, 3059–3066 (2016).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016).
Hubbard, B. P. & Sinclair, D. A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 35, 146–154 (2014).
Camandola, S. & Mattson, M. P. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 36, 1474–1492 (2017).
Fraser, J., Cabodevilla, A. G., Simpson, J. & Gammoh, N. Interplay of autophagy, receptor tyrosine kinase signalling and endocytic trafficking. Essays Biochem. 61, 597–607 (2017).
Wauson, E. M., Dbouk, H. A., Ghosh, A. B. & Cobb, M. H. G protein-coupled receptors and the regulation of autophagy. Trends Endocrinol. Metab. 25, 274–282 (2014).
Kondratskyi, A., Kondratska, K., Skryma, R., Klionsky, D. J. & Prevarskaya, N. Ion channels in the regulation of autophagy. Autophagy 14, 3–21 (2017).
Huang, Y., Todd, N. & Thathiah, A. The role of GPCRs in neurodegenerative diseases: avenues for therapeutic intervention. Curr. Opin. Pharmacol. 32, 96–110 (2017).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Pan, H., Yan, Y., Liu, C. & Finkel, T. The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy 13, 1235–1238 (2017).
Tsuboyama, K. et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 (2016).
Gowrishankar, S., Wu, Y. & Ferguson, S. M. Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J. Cell Biol. 216, 3291–3305 (2017).
Maday, S. Mechanisms of neuronal homeostasis: autophagy in the axon. Brain Res. 1649, 143–150 (2016).
Tammineni, P., Jeong, Y. Y., Feng, T., Aikal, D. & Cai, Q. Impaired axonal retrograde trafficking of the retromer complex augments lysosomal deficits in Alzheimer's disease neurons. Hum. Mol. Genet. 26, 4352–4366 (2017).
Berman, D. E., Ringe, D., Petsko, G. A. & Small, S. A. The use of pharmacological retromer chaperones in Alzheimer's disease and other endosomal-related disorders. Neurotherapeutics 12, 12–18 (2015).
Martens, S., Nakamura, S. & Yoshimori, T. Phospholipids in autophagosome formation and fusion. J. Mol. Biol. 428, 4819–4827 (2016).
Kaminskyy, V. & Zhivotovsky, B. Proteases in autophagy. Biochim. Biophys. Acta 1824, 44–50 (2012).
Colacurcio, D. J. & Nixon, R. A. Disorders of lysosomal acidification — The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88 (2016).
Mauvezin, C., Nagy, P., Juhasz, G. & Neufeld, T. P. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6, 7007 (2015).
Lu, S. & Nixon, R. A. in Lysosomes: Biology, Diseases, and Therapeutics 315–356 (John Wiley & Sons, 2016).
Platt, F. M. Emptying the stores: lysosomal diseases and therapeutic strategies. Nat. Rev. Drug Discov. 17, 133–150 (2017).
Settembre, C., Fraldi, A., Medina, D. L. & Ballabio, A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 (2013).
Xilouri, M. & Stefanis, L. Chaperone mediated autophagy to the rescue: A new-fangled target for the treatment of neurodegenerative diseases. Mol. Cell Neurosci. 66, 29–36 (2015).
Catarino, S., Pereira, P. & Girao, H. Molecular control of chaperone-mediated autophagy. Essays Biochem. 61, 663–674 (2017).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Medinas, D. B., Valenzuela, V. & Hetz, C. Proteostasis disturbance in amyotrophic lateral sclerosis. Hum. Mol. Genet. 26, R91–R104 (2017).
de Poot, S. A. H., Tian, G. & Finley, D. Meddling with fate: the proteasomal deubiquitinating enzymes. J. Mol. Biol. 429, 3525–3545 (2017).
Bonet-Costa, V., Pomatto, L. C. & Davies, K. J. The proteasome and oxidative stress in Alzheimer's disease. Antioxid. Redox Signal 25, 886–901 (2016).
Wrobel, L. et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488 (2015).
Hegde, A. N. Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 138, 98–110 (2017).
Wolfe, K. J., Ren, H. Y., Trepte, P. & Cyr, D. M. Polyglutamine-rich suppressors of huntingtin toxicity act upstream of Hsp70 and Sti1 in spatial quality control of amyloid-like proteins. PLoS One 9, e95914 (2014).
Menzies, F. M. et al. Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity. Cell Death Differ. 22, 433–444 (2015).
Ciechanover, A. & Kwon, Y. T. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp. Mol. Med. 47, e147 (2015).
Vilchez, D., Saez, I. & Dillin, A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014).
Mollereau, B. et al. Adaptive preconditioning in neurological diseases – therapeutic insights from proteostatic perturbations. Brain Res. 1648, 603–616 (2016).
Sorrentino, V. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552, 187–193 (2017).
Huang, Y. A., Zhou, B., Wernig, M. & Sudhof, T. C. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell 168, 427–441.e21 (2017).
Zlokovic, B. V. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol. 70, 440–444 (2013).
Zhang, Z., Xie, M. & Ye, K. Asparagine endopeptidase is an innovative therapeutic target for neurodegenerative diseases. Expert Opin. Ther. Targets 20, 1237–1245 (2016).
Simonovitch, S. et al. Impaired autophagy in APOE4 astrocytes. J. Alzheimers Dis. 51, 915–927 (2016).
Lee, J. H. et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 (2010).
Martin-Maestro, P. et al. Mitophagy failure in fibroblasts and iPSC-derived neurons of Alzheimer's disease-associated presenilin 1 mutation. Front. Mol. Neurosci. 10, 291 (2017).
Sannerud, R. et al. Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ pool. Cell 166, 193–208 (2016).
Lauritzen, I. et al. Intraneuronal aggregation of the β-CTF fragment of APP (C99) induces Aβ-independent lysosomal-autophagic pathology. Acta Neuropathol. 132, 257–276 (2016).
Seixas da Silva, G. S. et al. Amyloid-β oligomers transiently inhibit AMP-activated kinase and cause metabolic defects in hippocampal neurons. J. Biol. Chem. 292, 7395–7406 (2017).
Myeku, N. & Duff, K. Targeting the 26S proteasome to protect against proteotoxic diseases. Trends Mol. Med. 24, 15–29 (2018).
Corti, O., Lesage, S. & Brice, A. What genetics tells us about the cause and mechanisms of Parkinson's disease: Physiol. Rev. Transm. Suppl. 91, 1161–1128 (2011).
Youle, R. J. & Narendra, D. P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011).
Chen, Z. C. et al. Phosphorylation of amyloid precursor protein by mutant LRRK2 promotes AICD activity and neurotoxicity in Parkinson's disease. Sci. Signal. 10, eaam6790 (2017).
Giaime, E. et al. Age-dependent dopaminergic neurodegeneration and impairment of the autophagy-lysosomal pathway in LRRK-deficient mice. Neuron 96, 796–807.e6 (2017).
Manzoni, C. et al. mTOR independent regulation of macroautophagy by Leucine Rich Repeat Kinase 2 via Beclin-1. Sci. Rep. 6, 35106 (2016).
Aflaki, E., Westbroek, W. & Sidransky, E. The complicated relationship between Gaucher Disease and parkinsonism: insights from a rare disease. Neuron 93, 737–746 (2017).
Noelker, C. et al. Glucocerebrosidase deficiency and mitochondrial impairment in experimental Parkinson disease. J. Neurol. Sci. 356, 129–136 (2015).
Bento, C. F., Ashkenazi, A., Jimenez-Sanchez, M. & Rubinsztein, D. C. The Parkinson's disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat. Commun. 7, 11803 (2016).
Kong, S. M. et al. Parkinson's disease-linked human PARK9/ATP13A2 maintains zinc homeostasis and promotes α-synuclein externalization via exosomes. Hum. Mol. Genet. 23, 2816–2833 (2014).
Tsunemi, T. & Krainc, D. Zn2+ dyshomeostasis caused by loss of ATP13A2/PARK9 leads to lysosomal dysfunction and α-synuclein accumulation. Hum. Mol. Genet. 23, 2791–2801 (2014).
Zondler, L. et al. Proteasome impairment by α-synuclein. PLOS One 12, e0184040 (2017).
Sala, G., Marinig, D., Arosio, A. & Ferrarese, C. Role of chaperone-mediated autophagy dysfunctions in the pathogenesis of Parkinson's disease. Front. Mol. Neurosci. 9, 157 (2016).
Li, C. & Gotz, J. Tau-based therapies in neurodegeneration: opportunities and challenges. Nat. Rev. Drug Discov. 16, 863–883 (2017).
Gao, F. B., Almeida, S. & Lopez-Gonzalez, R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 36, 2931–2950 (2017).
Gotzl, J. K., Lang, C. M., Haass, C. & Capell, A. Impaired protein degradation in FTLD and related disorders. Ageing Res. Rev. 32, 122–139 (2016).
Ramesh, N. & Pandey, U. B. Autophagy dysregulation in ALS: when protein aggregates get out of hand. Front. Mol. Neurosci. 10, 263 (2017).
Guo, Q. et al. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705.e12 (2018).
Tanaka, Y. et al. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 26, 969–988 (2017).
Oakes, J. A., Davies, M. C. & Collins, M. O. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol. Brain 10, 5 (2017).
Nassif, M., Woehlbier, U. & Manque, P. A. The enigmatic role of C9ORF72 in autophagy. Front. Neurosci. 11, 442 (2017).
Ji, Y. J., Ugolino, J., Brady, N. R., Hamacher-Brady, A. & Wang, J. Systemic deregulation of autophagy upon loss of ALS- and FTD-linked C9orf72. Autophagy 13, 1254–1255 (2017).
Henriques, A. et al. Inhibition of β-glucocerebrosidase activity preserves motor unit integrity in a mouse model of amyotrophic lateral sclerosis. Sci. Rep. 7, 5235 (2017).
Lin, G., Mao, D. & Bellen, H. J. Amyotrophic lateral sclerosis pathogenesis converges on defects in protein homeostasis associated with TDP-43 mislocalization and proteasome-mediated degradation overload. Curr. Top. Dev. Biol. 121, 111–171 (2017).
Kaliszewski, M., Knott, A. B. & Bossy-Wetzel, E. Primary cilia and autophagic dysfunction in Huntington's disease. Cell Death Differ. 22, 1413–1424 (2015).
Mealer, R. G., Murray, A. J., Shahani, N., Subramaniam, S. & Snyder, S. H. Rhes, a striatal-selective protein implicated in Huntington disease, binds beclin-1 and activates autophagy. J. Biol. Chem. 289, 3547–3554 (2014).
Ashkenazi, A. et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 118–111 (2017).
Bauer, P. O. et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 28, 256–263 (2010).
Xilouri, M., Vogiatzi, T., Vekrellis, K., Park, D. & Stefanis, L. Abberant α-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One 4, e5515 (2009).
Her, L. S. et al. The differential profiling of ubiquitin-proteasome and autophagy systems in different tissues before the onset of Huntington's disease models. Brain Pathol. 25, 481–490 (2015).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Halliday, M. et al. Repurposed drugs targeting eIF2α-P-mediated translational repression prevent neurodegeneration in mice. Brain 140, 1768–1773 (2017).
Mogk, A., Bukau, B. & Kampinga, H. H. Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226 (2018).
Congdon, E. E. et al. Methylthioninium chloride (methylene blue) induces autophagy and attenuates tauopathy in vitro and in vivo. Autophagy 8, 609–622 (2012).
Xie, L. et al. Methylene blue induces macroautophagy through 5′ adenosine monophosphate-activated protein kinase pathway to protect neurons from serum deprivation. Front. Cell Neurosci. 7, 56 (2013).
Williams, A. et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 4, 295–305 (2008).
Rose, C. et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum. Mol. Genet. 19, 2144–2153 (2010).
Sarkar, S., Ravikumar, B., Floto, R. A. & Rubinsztein, D. C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16, 46–56 (2009).
Rao, M. V. et al. Specific calpain inhibition by calpastatin prevents tauopathy and neurodegeneration and restores normal lifespan in tau P301L mice. J. Neurosci. 34, 9222–9234 (2014).
Rao, M. V., Campbell, J., Palaniappan, A., Kumar, A. & Nixon, R. A. Calpastatin inhibits motor neuron death and increases survival of hSOD1(G93A) mice. J. Neurochem. 137, 253–265 (2016).
Park, S. Y. et al. Cilostazol modulates autophagic degradation of β-amyloid peptide via SIRT1-coupled LKB1/AMPKα signaling in neuronal cells. PLOS One 11, e0160620 (2016).
Ayasolla, K. R., Singh, A. K. & Singh, I. 5-Aminoimidazole-4-carboxamide-1-β-4-ribofuranoside (AICAR) attenuates the expression of LPS- and Aβ peptide-induced inflammatory mediators in astroglia. J. Neuroinflamm. 2, 21 (2005).
Dulovic, M. et al. The protective role of AMP-activated protein kinase in α-synuclein neurotoxicity in vitro. Neurobiol. Dis. 63, 1–11 (2014).
Walter, C. et al. Activation of AMPK-induced autophagy ameliorates Huntington disease pathology in vitro. Neuropharmacology 108, 24–38 (2016).
Zhang, Z. H. et al. Selenomethionine mitigates cognitive decline by targeting both Tau hyperphosphorylation and autophagic clearance in an Alzheimer's disease mouse model. J. Neurosci. 37, 2449–2462 (2017).
Park, S. J. et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433 (2012).
Vingtdeux, V. et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-β peptide metabolism. J. Biol. Chem. 285, 9100–9113 (2010).
Parker, J. A. et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet. 37, 349–350 (2005).
Martin-Montalvo, A. et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192 (2013).
Kickstein, E. et al. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc. Natl Acad. Sci. USA 107, 21830–21835 (2010).
Chen, B., Teng, Y., Zhang, X., Lv, X. & Yin, Y. Metformin alleviated Aβ-induced apoptosis via the suppression of JNK MAPK signaling pathway in cultured hippocampal neurons. Biomed. Res. Int. 2016, 1421430 (2016).
Patil, S. P., Jain, P. D., Ghumatkar, P. J., Tambe, R. & Sathaye, S. Neuroprotective effect of metformin in MPTP-induced Parkinson's disease in mice. Neuroscience 277, 747–754 (2014).
Castillo, K. et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9, 1308–1320 (2013).
Sarkar, S., Davies, J. E., Huang, Z., Tunnacliffe, A. & Rubinsztein, D. C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J. Biol. Chem. 282, 5641–5652 (2007).
Kruger, U., Wang, Y., Kumar, S. & Mandelkow, E. M. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging 33, 2291–2305 (2012).
Du, J., Liang, Y., Xu, F., Sun, B. & Wang, Z. Trehalose rescues Alzheimer's disease phenotypes in APP/PS1 transgenic mice. J. Pharm. Pharmacol. 65, 1753–1756 (2013).
Tanaka, M. et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 10, 148–154 (2004).
Schaeffer, V. & Goedert, M. Stimulation of autophagy is neuroprotective in a mouse model of human tauopathy. Autophagy 8, 1686–1687 (2012).
Sarkar, S. et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170, 1101–1111 (2005).
Shimada, K. et al. Long-term oral lithium treatment attenuates motor disturbance in tauopathy model mice: implications of autophagy promotion. Neurobiol. Dis. 46, 101–108 (2012).
Fornai, F. et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA 105, 2052–2057 (2008).
Li, H. et al. Biochemical protective effect of 1,25-dihydroxyvitamin D3 through autophagy induction in the MPTP mouse model of Parkinson's disease. Neuroreport 26, 669–674 (2015).
Webb, J. L., Ravikumar, B., Atkins, J., Skepper, J. N. & Rubinsztein, D. C. α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 (2003).
Ravikumar, B., Duden, R. & Rubinsztein, D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 (2002).
Ryu, H. H. et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol. Aging 35, 2822–2831 (2014).
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 (2004).
Wang, I. F., Tsai, K. J. & Shen, C. K. Autophagy activation ameliorates neuronal pathogenesis of FTLD-U mice: a new light for treatment of TARDBP/TDP-43 proteinopathies. Autophagy 9, 239–240 (2013).
Liu, K., Shi, N., Sun, Y., Zhang, T. & Sun, X. Therapeutic effects of rapamycin on MPTP-induced parkinsonism in mice. Neurochem. Res. 38, 201–207 (2013).
Caccamo, A., Majumder, S., Richardson, A., Strong, R. & Oddo, S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-β, and Tau: effects on cognitive impairments. J. Biol. Chem. 285, 13107–13120 (2010).
Jiang, T. et al. Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance. Neuropharmacology 85, 121–130 (2014).
Siracusa, R. et al. Neuroprotective effects of temsirolimus in animal models of Parkinson's disease. Mol. Neurobiol. 55, 2403–2419 (2017).
Menzies, F. M. et al. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain 133, 93–104 (2010).
Sarkar, S. et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 3, 331–338 (2007).
Satish Bollimpelli, V. & Kondapi, A. K. Differential sensitivity of immature and mature ventral mesencephalic neurons to rotenone induced neurotoxicity in vitro. Toxicol. In Vitro 30, 545–551 (2015).
Pandey, N., Strider, J., Nolan, W. C., Yan, S. X. & Galvin, J. E. Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol. 115, 479–489 (2008).
Jiang, T. F. et al. Curcumin ameliorates the neurodegenerative pathology in A53T α-synuclein cell model of Parkinson's disease through the downregulation of mTOR/p70S6K signaling and the recovery of macroautophagy. J. Neuroimmune Pharmacol. 8, 356–369 (2013).
Spinelli, K. J., Osterberg, V. R., Meshul, C. K., Soumyanath, A. & Unni, V. K. Curcumin treatment improves motor behavior in α-synuclein transgenic mice. PLoS One 10, e0128510 (2015).
Ma, Q. L. et al. Curcumin suppresses soluble tau dimers and corrects molecular chaperone, synaptic, and behavioral deficits in aged human tau transgenic mice. J. Biol. Chem. 288, 4056–4065 (2013).
Yang, F. et al. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 280, 5892–5901 (2005).
Medina, D. L. et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 (2015).
Xiao, Q. et al. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Aβ generation and amyloid plaque pathogenesis. J. Neurosci. 35, 12137–12151 (2015).
Kim, S. et al. Fisetin stimulates autophagic degradation of phosphorylated tau via the activation of TFEB and Nrf2 transcription factors. Sci. Rep. 6, 24933 (2016).
Hori, Y. et al. A Food and Drug Administration-approved asthma therapeutic agent impacts amyloid β in the brain in a transgenic model of Alzheimer disease. J. Biol. Chem. 290, 1966–1978 (2015).
Li, Y. et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 18, 1065–1077 (2016).
Schlatterer, S. D., Acker, C. M. & Davies, P. c-Abl in neurodegenerative disease. J. Mol. Neurosci. 45, 445–452 (2011).
Hebron, M. L., Lonskaya, I. & Moussa, C. E. Nilotinib reverses loss of dopamine neurons and improves motor behavior via autophagic degradation of α-synuclein in Parkinson's disease models. Hum. Mol. Genet. 22, 3315–3328 (2013).
Wenqiang, C. et al. Parkin-mediated reduction of nuclear and soluble TDP-43 reverses behavioral decline in symptomatic mice. Hum. Mol. Genet. 23, 4960–4969 (2014).
Pagan, F. et al. Nilotinib effects in Parkinson's disease and dementia with Lewy bodies. J. Parkinsons Dis. 6, 503–517 (2016).
Satoh, A., Imai, S. I. & Guarente, L. The brain, sirtuins, and ageing. Nat. Rev. Neurosci. 18, 362–374 (2017).
Kang, H. T. & Hwang, E. S. Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Aging Cell 8, 426–438 (2009).
Wu, M. F., Yin, J. H., Hwang, C. S., Tang, C. M. & Yang, D. I. NAD attenuates oxidative DNA damages induced by amyloid β-peptide in primary rat cortical neurons. Free Radic. Res. 48, 794–805 (2014).
Liu, D. et al. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. Neurobiol. Aging 34, 1564–1580 (2013).
Deng, H. & Mi, M. T. Resveratrol attenuates Aβ25-35 caused neurotoxicity by inducing autophagy through the TyrRS-PARP1-SIRT1 signaling pathway. Neurochem. Res. 41, 2367–2379 (2016).
Martire, S. et al. Bioenergetic impairment in animal and cellular models of Alzheimer's disease: PARP-1 inhibition rescues metabolic dysfunctions. J. Alzheimers Dis. 54, 307–324 (2016).
Park, S. H. et al. Protective effect of the phosphodiesterase III inhibitor cilostazol on amyloid β-induced cognitive deficits associated with decreased amyloid β accumulation. Biochem. Biophys. Res. Commun. 408, 602–608 (2011).
Lee, H. R. et al. Attenuation of β-amyloid-induced tauopathy via activation of CK2α/SIRT1: targeting for cilostazol. J. Neurosci. Res. 92, 206–217 (2014).
Madeo, F., Pietrocola, F., Eisenberg, T. & Kroemer, G. Caloric restriction mimetics: towards a molecular definition. Nat. Rev. Drug Discov. 13, 727–740 (2014).
Millan, M. J. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer's disease: an integrative review. Prog. Neurobiol. 156, 1–68 (2017).
Park, G. et al. Regulation of histone acetylation by autophagy in Parkinson disease. J. Biol. Chem. 291, 3531–3540 (2016).
Madeo, F., Eisenberg, T., Pietrocola, F. & Kroemer, G. Spermidine in health and disease. Science 359, eaan2788 (2018).
Yang, Y. et al. Induction of autophagy by spermidine is neuroprotective via inhibition of caspase 3-mediated Beclin 1 cleavage. Cell Death Dis. 8, e2738 (2017).
Wang, I. F. et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc. Natl Acad. Sci. USA 109, 15024–15029 (2012).
Buttner, S. et al. Spermidine protects against α-synuclein neurotoxicity. Cell Cycle 13, 3903–3908 (2014).
Marino, G. et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 53, 710–725 (2014).
Aubry, S. et al. Assembly and interrogation of Alzheimer's disease genetic networks reveal novel regulators of progression. PLoS One 10, e0120352 (2015).
Yao, J. et al. Neuroprotection by cyclodextrin in cell and mouse models of Alzheimer disease. J. Exp. Med. 209, 2501–2513 (2012).
Shoji-Kawata, S. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013).
Lu, J. H. et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of α-synuclein in neuronal cells via inducing autophagy. Autophagy 8, 98–108 (2012).
Di Rita, A. & Strappazzon, F. AMBRA1, a novel BH3-like protein: new insights into the AMBRA1-BCL2-family proteins relationship. Int. Rev. Cell. Mol. Biol. 330, 85–113 (2017).
Pedro, J. M. et al. BAX and BAK1 are dispensable for ABT-737-induced dissociation of the BCL2-BECN1 complex and autophagy. Autophagy 11, 452–459 (2015).
Rocchi, A. et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer's disease. PLoS Genet. 13, e1006962 (2017).
Salminen, A. et al. Impaired autophagy and APP processing in Alzheimer's disease: the potential role of Beclin 1 interactome. Prog. Neurobiol. 106–107, 33–54 (2013).
Vidoni, C., Secomandi, E., Castiglioni, A., Melone, M. A. B. & Isidoro, C. Resveratrol protects neuronal-like cells expressing mutant Huntingtin from dopamine toxicity by rescuing ATG4-mediated autophagosome formation. Neurochem. Int. 117, 174–187 (2017).
Kovacs, T. et al. The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci. Rep. 7, 42014 (2017).
Seyb, K. I., Ansar, S., Bean, J. & Michaelis, M. L. β-Amyloid and endoplasmic reticulum stress responses in primary neurons: effects of drugs that interact with the cytoskeleton. J. Mol. Neurosci. 28, 111–123 (2006).
Zhang, B. et al. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J. Neurosci. 32, 3601–3611 (2012).
Kast, D. J. & Dominguez, R. The Cytoskeleton-autophagy connection. Curr. Biol. 27, R318–R326 (2017).
Coutts, A. S. & La Thangue, N. B. Regulation of actin nucleation and autophagosome formation. Cell. Mol. Life Sci. 73, 3249–3263 (2016).
Wang, Z. et al. The Vici syndrome protein EPG5 Is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell 63, 781–795 (2016).
Shi, Y. et al. Haploinsufficiency leads to neuro-degeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 24, 313–325 (2018).
Mecozzi, V. J. et al. Pharmacological chaperones stabilize retromer to limit APP processing. Nat. Chem. Biol. 10, 443–449 (2014).
Coffey, E. E., Beckel, J. M., Laties, A. M. & Mitchell, C. H. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer's disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263, 111–124 (2014).
Moruno-Manchon, J. F. et al. TFEB ameliorates the impairment of the autophagy-lysosome pathway in neurons induced by doxorubicin. Aging 8, 3507–3519 (2016).
Wang, W. et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl Acad. Sci. USA 112, E1373–1381 (2015).
Bae, M. et al. Activation of TRPML1 clears intraneuronal Aβ in preclinical models of HIV infection. J. Neurosci. 34, 11485–11503 (2014).
Kao, A. W., McKay, A., Singh, P. P., Brunet, A. & Huang, E. J. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18, 325–333 (2017).
Arrant, A. E., Onyilo, V. C., Unger, D. E. & Roberson, E. D. Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J. Neurosci. 38, 2341–2358 (2018).
Kilpatrick, K., Zeng, Y., Hancock, T. & Segatori, L. Genetic and chemical activation of TFEB mediates clearance of aggregated α-synuclein. PLoS One 10, e0120819 (2015).
Seo, B. R., Lee, S. J., Cho, K. S., Yoon, Y. H. & Koh, J. Y. The zinc ionophore clioquinol reverses autophagy arrest in chloroquine-treated ARPE-19 cells and in APP/mutant presenilin-1-transfected Chinese hamster ovary cells. Neurobiol. Aging 36, 3228–3238 (2015).
Cherny, R. A. et al. Treatment with a copper-zinc chelator markedly and rapidly inhibits β-amyloid accumulation in Alzheimer's disease transgenic mice. Neuron 30, 665–676 (2001).
Sun, B. et al. Cystatin C-cathepsin B axis regulates amyloid β levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron 60, 247–257 (2008).
Sardi, S. P. et al. Glucosylceramide synthase inhibition alleviates aberrations in synucleinopathy models. Proc. Natl Acad. Sci. USA 114, 2699–2704 (2017).
Yang, S. Y., Beavan, M., Chau, K. Y., Taanman, J. W. & Schapira, A. H. A. Human neural crest stem cell-derived dopaminergic neuronal model recapitulates biochemical abnormalities in GBA1 mutation carriers. Stem Cell Rep. 8, 728–742 (2017).
Sanchez-Martinez, A. et al. Parkinson disease-linked GBA mutation effects reversed by molecular chaperones in human cell and fly models. Sci. Rep. 6, 31380 (2016).
Migdalska-Richards, A., Daly, L., Bezard, E. & Schapira, A. H. Ambroxol effects in glucocerebrosidase and α-synuclein transgenic mice. Ann. Neurol. 80, 766–775 (2016).
Aflaki, E. et al. A new glucocerebrosidase chaperone reduces α-synuclein and glycolipid levels in iPSC-derived dopaminergic neurons from patients with Gaucher disease and parkinsonism. J. Neurosci. 36, 7441–7452 (2016).
Song, W., Wang, F., Lotfi, P., Sardiello, M. & Segatori, L. 2-Hydroxypropyl-β-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J. Biol. Chem. 289, 10211–10222 (2014).
Duan, W. J. et al. A SIRT3/AMPK/autophagy network orchestrates the protective effects of trans-resveratrol in stressed peritoneal macrophages and RAW 264.7 macrophages. Free Radic. Biol. Med. 95, 230–242 (2016).
Writing, G. & Edaravone, A. L. S. S. G. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet. Neurol. 16, 505–512 (2017).
Li, S. & Laher, I. Exercise pills: at the starting line. Trends Pharmacol. Sci. 36, 906–917 (2015).
Lackie, R. E. et al. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front. Neurosci. 11, 254 (2017).
Neef, D. W., Jaeger, A. M. & Thiele, D. J. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 10, 930–944 (2011).
Kalmar, B., Lu, C. H. & Greensmith, L. The role of heat shock proteins in Amyotrophic Lateral Sclerosis: the therapeutic potential of Arimoclomol. Pharmacol. Ther. 141, 40–54 (2014).
Kalmar, B. & Greensmith, L. Activation of the heat shock response in a primary cellular model of motoneuron neurodegeneration-evidence for neuroprotective and neurotoxic effects. Cell. Mol. Biol. Lett. 14, 319–335 (2009).
Kieran, D. et al. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat. Med. 10, 402–405 (2004).
Wang, A. M. et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 9, 112–118 (2013).
Lee, J. H. et al. Facilitated Tau degradation by USP14 aptamers via enhanced proteasome activity. Sci. Rep. 5, 10757 (2015).
Kiprowska, M. J. et al. Neurotoxic mechanisms by which the USP14 inhibitor IU1 depletes ubiquitinated proteins and Tau in rat cerebral cortical neurons: relevance to Alzheimer's disease. Biochim. Biophys. Acta 1863, 1157–1170 (2017).
Boselli, M. et al. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J. Biol. Chem. 292, 19209–19221 (2017).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Wang, B. et al. A CNS-permeable Hsp90 inhibitor rescues synaptic dysfunction and memory loss in APP-overexpressing Alzheimer's mouse model via an HSF1-mediated mechanism. Mol. Psychiatry 22, 990–1001 (2017).
Petrucelli, L. et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 13, 703–714 (2004).
Danzer, K. M. et al. Heat-shock protein 70 modulates toxic extracellular α-synuclein oligomers and rescues trans-synaptic toxicity. FASEB J. 25, 326–336 (2011).
Auluck, P. K. & Bonini, N. M. Pharmacological prevention of Parkinson disease in Drosophila. Nat. Med. 8, 1185–1186 (2002).
Agrawal, N. et al. Identification of combinatorial drug regimens for treatment of Huntington's disease using Drosophila. Proc. Natl Acad. Sci. USA 102, 3777–3781 (2005).
Chen, Y. et al. Hsp90 chaperone inhibitor 17-AAG attenuates Aβ-induced synaptic toxicity and memory impairment. J. Neurosci. 34, 2464–2470 (2014).
Ho, S. W. et al. Effects of 17-allylamino-17-demethoxygeldanamycin (17-AAG) in transgenic mouse models of frontotemporal lobar degeneration and Alzheimer's disease. Transl Neurodegener. 2, 24 (2013).
Labbadia, J. et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J. Clin. Invest. 121, 3306–3319 (2011).
Guo, X., Huang, X. & Chen, M. J. Reversible phosphorylation of the 26S proteasome. Protein Cell 8, 255–272 (2017).
VerPlank, J. J. S. & Goldberg, A. L. Regulating protein breakdown through proteasome phosphorylation. Biochem. J. 474, 3355–3371 (2017).
Bate, C. & Williams, A. cAMP-Inhibits cytoplasmic phospholipase A2 and protects neurons against amyloid-β-induced synapse damage. Biol. (Basel) 4, 591–606 (2015).
Myeku, N. et al. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22, 46–53 (2016).
Lokireddy, S., Kukushkin, N. V. & Goldberg, A. L. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc. Natl Acad. Sci. USA 112, E7176–E7185 (2015).
Lin, J. T. et al. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington's disease. Mol. Cell. Biol. 33, 1073–1084 (2013).
Djakovic, S. N. et al. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J. Neurosci. 32, 5126–5131 (2012).
Leestemaker, Y. et al. Proteasome activation by small molecules. Cell Chem. Biol. 24, 725–736.e7 (2017).
Crew, A. P. et al. Identification and characterization of von hippel-lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J. Med. Chem. 61, 583–598 (2017).
Collins, I., Wang, H., Caldwell, J. J. & Chopra, R. Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway. Biochem. J. 474, 1127–1147 (2017).
Chu, T. T. et al. Specific knockdown of endogenous tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem. Biol. 23, 453–461 (2016).
Clift, D. et al. A Method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706 (2017).
Choi, J. S. et al. cIAPs promote the proteasomal degradation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 480, 422–428 (2016).
Vangala, J. R., Sotzny, F., Kruger, E., Deshaies, R. J. & Radhakrishnan, S. K. Nrf1 can be processed and activated in a proteasome-independent manner. Curr. Biol. 26, R834–R835 (2016).
Pajares, M., Cuadrado, A. & Rojo, A. I. Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases. Redox Biol. 11, 543–553 (2017).
Tsakiri, E. N. et al. Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell 12, 802–813 (2013).
Opattova, A., Cente, M., Novak, M. & Filipcik, P. The ubiquitin proteasome system as a potential therapeutic target for treatment of neurodegenerative diseases. Gen. Physiol. Biophys. 34, 337–352 (2015).
Rousseau, A. & Bertolotti, A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189 (2016).
Arias, E. et al. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell 59, 270–284 (2015).
Anguiano, J. et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 9, 374–382 (2013).
Lopez, A. et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebrafish model by autophagy induction. Brain 140, 1128–1146 (2017).
Hou, Y. S. et al. Sestrin2 protects dopaminergic cells against rotenone toxicity through ampk-dependent autophagy activation. Mol. Cell. Biol. 35, 2740–2751 (2015).
Chen, Y. S., Chen, S. D., Wu, C. L., Huang, S. S. & Yang, D. I. Induction of sestrin2 as an endogenous protective mechanism against amyloid β-peptide neurotoxicity in primary cortical culture. Exp. Neurol. 253, 63–71 (2014).
Shafiei, S. S., Guerrero-Munoz, M. J. & Castillo-Carranza, D. L. Tau oligomers: cytotoxicity, propagation, and mitochondrial damage. Front. Aging Neurosci. 9, 83 (2017).
Valdinocci, D., Radford, R. A., Siow, S. M., Chung, R. S. & Pountney, D. L. Potential modes of intercellular α-synuclein transmission. Int. J. Mol. Sci. 18, E469 (2017).
Laulagnier, K. et al. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell. Mol. Life Sci. 75, 757–773 (2017).
Wu, J. W. et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19, 1085–1092 (2016).
Jha, N. K. et al. Impact of insulin degrading enzyme and neprilysin in Alzheimer's disease biology: characterization of putative cognates for therapeutic applications. J. Alzheimers Dis. 48, 891–917 (2015).
Baranello, R. J. et al. Amyloid-β protein clearance and degradation (ABCD) pathways and their role in Alzheimer's disease. Curr. Alzheimer Res. 12, 32–46 (2015).
Ruan, L. et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446 (2017).
Saido, T. & Leissring, M. A. Proteolytic degradation of amyloid β-protein. Cold Spring Harb. Perspect. Med. 2, a006379 (2012).
Miller, J. P. et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron 67, 199–212 (2010).
Brkic, M., Balusu, S., Libert, C. & Vandenbroucke, R. E. Friends or foes: matrix metalloproteinases and their multifaceted roles in neurodegenerative diseases. Mediators Inflamm. 2015, 620581 (2015).
Kurochkin, I. V., Guarnera, E. & Berezovsky, I. N. Insulin-degrading enzyme in the fight against Alzheimer's disease. Trends Pharmacol. Sci. 39, 49–58 (2017).
Maetzler, W. et al. Neprilysin activity in cerebrospinal fluid is associated with dementia and amyloid-β42 levels in Lewy body disease. J. Alzheimers Dis. 22, 933–938 (2010).
Jacobsen, J. S. et al. Enhanced clearance of Aβ in brain by sustaining the plasmin proteolysis cascade. Proc. Natl Acad. Sci. USA 105, 8754–8759 (2008).
Kim, K. S. et al. Proteolytic cleavage of extracellular α-synuclein by plasmin: implications for Parkinson disease. J. Biol. Chem. 287, 24862–24872 (2012).
Saito, S. & Ihara, M. New therapeutic approaches for Alzheimer's disease and cerebral amyloid angiopathy. Front. Aging Neurosci. 6, 290 (2014).
Spencer, B. et al. Lentivirus mediated delivery of neurosin promotes clearance of wild-type α-synuclein and reduces the pathology in an α-synuclein model of LBD. Mol. Ther. 21, 31–41 (2013).
Tarasoff-Conway, J. M. et al. Clearance systems in the brain-implications for Alzheimer disease. Nat. Rev. Neurol. 11, 457–470 (2015).
Drouin-Ouellet, J. et al. Cerebrovascular and blood-brain barrier impairments in Huntington's disease: potential implications for its pathophysiology. Ann. Neurol. 78, 160–177 (2015).
Cabezas, R. et al. Astrocytic modulation of blood brain barrier: perspectives on Parkinson's disease. Front. Cell Neurosci. 8, 211 (2014).
Shi, M. et al. CNS tau efflux via exosomes is likely increased in Parkinson's disease but not in Alzheimer's disease. Alzheimers Dement. 12, 1125–1131 (2016).
Sun, B. L. et al. Lymphatic drainage system of the brain: A novel target for intervention of neurological diseases. Prog Neurobiol. 163–164, 118–143 (2017).
Zlokovic, B. V. Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat. Rev. Neurosci. 12, 723–738 (2011).
Kanekiyo, T. & Bu, G. The low-density lipoprotein receptor-related protein 1 and amyloid-β clearance in Alzheimer's disease. Front. Aging Neurosci. 6, 93 (2014).
Ueno, M. et al. Blood-brain barrier and blood-cerebrospinal fluid barrier in normal and pathological conditions. Brain Tumor Pathol. 33, 89–96 (2016).
Bartels, A. L. Blood-brain barrier P-glycoprotein function in neurodegenerative disease. Curr. Pharm. Des. 17, 2771–2777 (2011).
Xie, L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013).
Iliff, J. J. et al. Impairment of glymphatic pathway function promotes tau pathology after traumatic brain injury. J. Neurosci. 34, 16180–16193 (2014).
Lan, Y. L. et al. Aquaporin 4 in astrocytes is a target for therapy in Alzheimer's disease. Curr. Pharm. Des. 23, 4948–4957 (2017).
Hoshi, A. et al. Expression of aquaporin 1 and aquaporin 4 in the temporal neocortex of patients with Parkinson's disease. Brain Pathol. 27, 160–168 (2017).
Xu, Z. et al. Deletion of aquaporin-4 in APP/PS1 mice exacerbates brain Aβ accumulation and memory deficits. Mol. Neurodegener. 10, 58 (2015).
Jiang, H. et al. RBD and neurodegenerative diseases. Mol. Neurobiol. 54, 2997–3006 (2017).
Yamamoto, N. et al. Epigallocatechin gallate induces extracellular degradation of amyloid β-protein by increasing neprilysin secretion from astrocytes through activation of ERK and PI3K pathways. Neuroscience 362, 70–78 (2017).
Van Kampen, J. M. & Kay, D. G. Progranulin gene delivery reduces plaque burden and synaptic atrophy in a mouse model of Alzheimer's disease. PLoS One 12, e0182896 (2017).
Bi Oh, S., Suh, N., Kim, I. & Lee, J. Y. Impacts of aging and amyloid-β deposition on plasminogen activators and plasminogen activator inhibitor-1 in the Tg2576 mouse model of Alzheimer's disease. Brain Res. 1597, 159–167 (2015).
Nalivaeva, N. N., Belyaev, N. D., Zhuravin, I. A. & Turner, A. J. The Alzheimer's amyloid-degrading peptidase, neprilysin: can we control it? Int. J. Alzheimers Dis. 2012, 383796 (2012).
Spampinato, S. F., Merlo, S., Sano, Y., Kanda, T. & Sortino, M. A. Astrocytes contribute to Aβ-induced blood-brain barrier damage through activation of endothelial MMP9. J. Neurochem. 142, 464–477 (2017).
Kingwell, K. Zeroing in on neurodegenerative α-synuclein. Nat. Rev. Drug Discov. 16, 371–373 (2017).
Wes, P. D., Sayed, F. A., Bard, F. & Gan, L. Targeting microglia for the treatment of Alzheimer's Disease. Glia 64, 1710–1732 (2016).
Sevigny, J. et al. The antibody aducanumab reduces Aβ plaques in Alzheimer's disease. Nature 537, 50–56 (2016).
Panza, F. et al. Tau-based therapeutics for Alzheimer's disease: active and passive immunotherapy. Immunotherapy 8, 1119–1134 (2016).
Schenk, D. B. et al. First-in-human assessment of PRX002, an anti-α-synuclein monoclonal antibody, in healthy volunteers. Mov Disord. 32, 211–218 (2017).
Shackleton, B., Crawford, F. & Bachmeier, C. Inhibition of ADAM10 promotes the clearance of Aβ across the BBB by reducing LRP1 ectodomain shedding. Fluids Barriers CNS 13, 14 (2016).
Shinohara, M. et al. Reduction of brain β-amyloid (Aβ) by fluvastatin, a hydroxymethylglutaryl-CoA reductase inhibitor, through increase in degradation of amyloid precursor protein C-terminal fragments (APP-CTFs) and Aβ clearance. J. Biol. Chem. 285, 22091–22102 (2010).
Qosa, H., Abuznait, A. H., Hill, R. A. & Kaddoumi, A. Enhanced brain amyloid-β clearance by rifampicin and caffeine as a possible protective mechanism against Alzheimer's disease. J. Alzheimers Dis. 31, 151–165 (2012).
Umeda, T. et al. Rifampicin is a candidate preventive medicine against amyloid-β and tau oligomers. Brain 139, 1568–1586 (2016).
Wan, W. et al. Aβ(1–42) oligomer-induced leakage in an in vitro blood-brain barrier model is associated with up-regulation of RAGE and metalloproteinases, and down-regulation of tight junction scaffold proteins. J. Neurochem. 134, 382–393 (2015).
Deane, R. et al. A multimodal RAGE-specific inhibitor reduces amyloid β-mediated brain disorder in a mouse model of Alzheimer disease. J. Clin. Invest. 122, 1377–1392 (2012).
Burstein, A. H. et al. Effect of TTP488 in patients with mild to moderate Alzheimer's disease. BMC Neurol. 14, 12 (2014).
Zhao, H. F. et al. Resveratrol decreases the insoluble Aβ1-42 level in hippocampus and protects the integrity of the blood-brain barrier in AD rats. Neuroscience 310, 641–649 (2015).
Corona, A. W., Kodoma, N., Casali, B. T. & Landreth, G. E. ABCA1 is necessary for bexarotene-mediated clearance of soluble amyloid β from the hippocampus of APP/PS1 mice. J. Neuroimmune Pharmacol. 11, 61–72 (2016).
Fan, C. H., Lin, C. Y., Liu, H. L. & Yeh, C. K. Ultrasound targeted CNS gene delivery for Parkinson's disease treatment. J. Control Release 261, 246–262 (2017).
Burgess, A., Huang, Y., Querbes, W., Sah, D. W. & Hynynen, K. Focused ultrasound for targeted delivery of siRNA and efficient knockdown of Htt expression. J. Control Release 163, 125–129 (2012).
McMahon, D., Bendayan, R. & Hynynen, K. Acute effects of focused ultrasound-induced increases in blood-brain barrier permeability on rat microvascular transcriptome. Sci. Rep. 7, 45657 (2017).
Maki, T. et al. Phosphodiesterase III inhibitor promotes drainage of cerebrovascular β-amyloid. Ann. Clin. Transl Neurol. 1, 519–533 (2014).
Ihara, M. et al. Cilostazol add-on therapy in patients with mild dementia receiving donepezil: a retrospective study. PLoS One 9, e89516 (2014).
Lundgaard, I. et al. Beneficial effects of low alcohol exposure, but adverse effects of high alcohol intake on glymphatic function. Sci. Rep. 8, 2246 (2018).
Jin, W. S. et al. Peritoneal dialysis reduces amyloid-β plasma levels in humans and attenuates Alzheimer-associated phenotypes in an APP/PS1 mouse model. Acta Neuropathol. 134, 207–220 (2017).
Domise, M. & Vingtdeux, V. AMPK in neurodegenerative diseases. EXS 107, 153–177 (2016).
Millan, M. J. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology 68, 2–82 (2013).
Smith, A. J. & Verkman, A. S. The “glymphatic” mechanism for solute clearance in Alzheimer's disease: game changer or unproven speculation? FASEB J. 32, 543–551 (2017).
Gonzalez-Marrero, I. et al. Choroid plexus dysfunction impairs β-amyloid clearance in a triple transgenic mouse model of Alzheimer's disease. Front. Cell Neurosci. 9, 17 (2015).
Alvira-Botero, X. & Carro, E. M. Clearance of amyloid-β peptide across the choroid plexus in Alzheimer's disease. Curr. Aging Sci. 3, 219–229 (2010).
Jeromin, A. & Bowser, R. Biomarkers in neurodegenerative diseases. Adv. Neurobiol. 15, 491–528 (2017).
Nguyen, C. T. O. et al. Retinal biomarkers provide “insight” into cortical pharmacology and disease. Pharmacol. Ther. 175, 151–177 (2017).
Nascimento-Ferreira, I. et al. Beclin 1 mitigates motor and neuropathological deficits in genetic mouse models of Machado-Joseph disease. Brain 136, 2173–2188 (2013).
Onofre, I. et al. Fibroblasts of Machado Joseph Disease patients reveal autophagy impairment. Sci. Rep. 6, 28220 (2016).
Song, P. et al. Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson's disease. Protein Cell 7, 114–129 (2016).
Martin-Jimenez, R., Campanella, M. & Russell, C. New zebrafish models of neurodegeneration. Curr. Neurol. Neurosci. Rep. 15, 33 (2015).
Zhang, Y. et al. Rescue of Pink1 deficiency by stress-dependent activation of autophagy. Cell Chem. Biol. 24, 471–480 (2017).
Wang, T., Lao, U. & Edgar, B. A. TOR-mediated autophagy regulates cell death in Drosophila neuro-degenerative disease. J. Cell Biol. 186, 703–711 (2009).
Hewitt, V. L. & Whitworth, A. J. Mechanisms of Parkinson's disease: lessons from Drosophila. Curr. Top. Dev. Biol. 121, 173–200 (2017).
Miyake, S., Takihara, Y., Yokota, S., Takamura, Y. & Inatani, M. Effect of microtubule disruption on dynamics of acidic organelles in the axons of primary cultured retinal ganglion cells. Curr. Eye Res. 43, 77–83 (2017).
Fouillet, A. et al. ER stress inhibits neuronal death by promoting autophagy. Autophagy 8, 915–926 (2012).
Palikaras, K., Daskalaki, I., Markaki, M. & Tavernarakis, N. Mitophagy and age-related pathologies: Development of new therapeutics by targeting mitochondrial turnover. Pharmacol. Ther. 178, 157–174 (2017).
Martinez-Vicente, M. Neuronal mitophagy in neurodegenerative diseases. Front. Mol. Neurosci. 10, 64 (2017).
Ashrafi, G., Schlehe, J. S., LaVoie, M. J. & Schwarz, T. L. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206, 655–670 (2014).
Du, F. et al. PINK1 signalling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer's disease. Brain 140, 3233–3251 (2017).
Di Maio, R. et al. α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Sci. Transl Med. 8, 342ra78 (2016).
Georgakopoulos, N. D., Wells, G. & Campanella, M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 13, 136–146 (2017).
Hertz, N. T. et al. A neo-substrate that amplifies catalytic activity of parkinson's-disease-related kinase PINK1. Cell 154, 737–747 (2013).
Hasson, S. A. et al. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem. Biol. 10, 1188–1197 (2015).
Gersch, M. et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 24, 920–930 (2017).
Dutta, D. et al. EphrinA2 regulates clathrin mediated KSHV endocytosis in fibroblast cells by coordinating integrin-associated signaling and c-Cbl directed polyubiquitination. PLoS Pathog. 9, e1003510 (2013).
East, D. A. et al. PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem. Biol. 21, 1585–1596 (2014).
Jang, S. Y., Kang, H. T. & Hwang, E. S. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J. Biol. Chem. 287, 19304–19314 (2012).
Wu, B. et al. Succinate-induced neuronal mitochondrial fission and hexokinase II malfunction in ischemic stroke: therapeutical effects of kaempferol. Biochim. Biophys. Acta 1863, 2307–2318 (2017).
Ryu, D. et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 22, 879–888 (2016).
Hylin, M. J. et al. A role for autophagy in long-term spatial memory formation in male rodents. J. Neurosci. Res. 96, 416–426 (2018).
Lee, J. H. & Lee, M. J. Isolation and characterization of RNA aptamers against a proteasome-associated deubiquitylating enzyme UCH37. Chembiochem. 18, 171–175 (2017).
Kim, J. H. et al. Inhibitory RNA aptamers of Tau oligomerization and their neuroprotective roles against proteotoxic stress. Mol. Pharmaceut. 13, 2039–2048 (2016).
Tsukakoshi, K., Abe, K., Sode, K. & Ikebukuro, K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 84, 5542–5547 (2012).
Wang, H. et al. MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson's disease by targeting to Bim. Brain Pathol. 26, 167–176 (2016).
Schoch, K. M. & Miller, T. M. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron 94, 1056–1070 (2017).
Lai, C. C. & Crews, C. M. Induced protein degradation: an emerging drug paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Wurz, R. P. et al. A “click chemistry platform” for the rapid synthesis of bispecific molecules for inducing protein degradation. J. Med. Chem. 61, 453–461 (2017).
Bourdenx, M. et al. Nanoparticles restore lysosomal acidification defects: implications for Parkinson and other lysosomal-related diseases. Autophagy 12, 472–483 (2016).
Gambaryan, P. Y., Kondrasheva, I. G., Severin, E. S., Guseva, A. A. & Kamensky, A. A. Increasing the efficiency of Parkinson's disease treatment using a poly(lactic-co-glycolic acid) (PLGA) based L-DOPA delivery system. Exp. Neurobiol. 23, 246–252 (2014).
Popp, L. & Segatori, L. Differential autophagic responses to nano-sized materials. Curr. Opin. Biotechnol. 36, 129–136 (2015).
Hernandez, D. et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 74, 277–284 (2012).
Han, Y., Khodr, C. E., Sapru, M. K., Pedapati, J. & Bohn, M. C. A. microRNA embedded AAV α-synuclein gene silencing vector for dopaminergic neurons. Brain Res. 1386, 15–24 (2011).
Carpentier, A. et al. Clinical trial of blood-brain barrier disruption by pulsed ultrasound. Sci. Transl Med. 8, 343re2 (2016).
Jeon, J., Kim, W., Jang, J., Isacson, O. & Seo, H. Gene therapy by proteasome activator, PA28γ, improves motor coordination and proteasome function in Huntington's disease YAC128 mice. Neuroscience 324, 20–28 (2016).
Decressac, M. et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl Acad. Sci. USA 110, E1817–1826 (2013).
Iaccarino, H. F. et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235 (2016).
Acknowledgements
The authors would like to thank S.-M. Rivet for help in the preparation of the figures, K. Duff, R. Jeggo, M.-C. Potier and C. Mannoury la Cour for helpful comments on the manuscript and M. Galliot and her colleagues in the Institut de recherche Servier (IDRS) Documentation Department for provision of papers relevant to this article. This article is based upon a small, focused symposium that was supported by an unrestricted grant from Advances in Neuroscience for Medical Innovation, which is affiliated with the Institut de Recherche Servier.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.J.M. is a full-time employee of Servier Pharmaceuticals and has no other competing interests to declare. E.B. has an equity stake in Motac Holding Ltd. and receives consultancy payments from Motac Neuroscience Ltd. D.C.R. is a consultant for E3Bio and has consulted for GlaxoSmithKline, AbbVie and AstraZeneca. He has received grant support from AbbVie and AstraZeneca. B.L. is a scientific founder of Casma Therapeutics. A.H. is a full-time employee of Neuro-Sys and has no other competing interests to declare. C.L. is a full-time employee of Servier Pharmaceuticals and has no other competing interests to declare. M.S. is a consultant for Servier Pharmaceuticals and President of Spedding Research Solutions SAS, a company performing research on potential therapies for amyotrophic lateral sclerosis. The author others declare no competing interests.
Supplementary information
Supplementary information
Supplementary Text (PDF 1794 kb)
Glossary
- Neurodegenerative disorders of ageing
-
(NDAs). A suite of neurodegenerative diseases including Alzheimer disease, Parkinson disease, amyotrophic lateral sclerosis and frontotemporal dementia that typically are diagnosed in elderly individuals. Most cases are sporadic, but rare forms are associated with mutations (Table 1). Huntington disease is an exception in being purely genetic and having a somewhat earlier onset at 30–50 years of age.
- Proteinopathies
-
A general term for disorders characterized by the build-up of excess, anomalously marked, misfolded and/or aggregated neurotoxic proteins like Aβ, tau or α-synuclein.
- Amyloid-β
-
(Aβ). The major neurotoxic product of amyloid precursor protein (APP) processing, including Aβ fragment 42 (Aβ42), which deposits into extracellular plaques in Alzheimer disease. It is toxic as oligomers and protofibrils because it, for example, disrupts synaptic transmission, damages mitochondria and impedes proteasomal clearance.
- Tau
-
A protein that stabilizes axonal microtubules. It is prone to cleavage, hyperphosphorylation and other modifications that trigger and/or follow microtubule dissociation. This leads to misfolding, oligomerization, synaptic mislocalization and interneuronal spreading. Aggregates, fibrils and intracellular neurofibrillary tangles are also formed.
- α-Synuclein
-
A phospholipid-binding protein abundant in presynaptic terminals and involved in the release and regulation of synaptic vesicles. α-Synuclein is a major component of Lewy bodies (protein and lipid aggregates) in Parkinson disease. Its spread and accumulation in dopaminergic cell bodies and other cell types are typical features of the disease.
- TAR DNA-binding protein 43
-
(TDP43). A normally nuclear protein that is associated with frontotemporal dementia and amyotrophic lateral sclerosis. In these diseases, it is found in the cytoplasm, where it aggregates.
- Glymphatic system
-
A system that serves as a cerebrospinal fluid-driven mechanism for flushing extracellular pools of neurotoxic protein into the circulation; it involves perivascular drainage, astrocytes and the lymph system.
- Blood–brain barrier
-
A physical and functional barrier that isolates the brain from the rest of the body. Certain nutrients, lipid vesicles and small molecules can enter, yet it excludes toxic elements that may damage the brain. It also ejects neurotoxic proteins and other unwanted material. Active transfer of neurotoxic proteins from the brain to the periphery involves specific classes of receptor and transporter.
- Lysosomes
-
Acidic compartments for the degradation of proteins and other cellular constituents. Substrate breakdown yields products such as amino acids, sugars and lipids, which are recycled.
- Aggresomes
-
Microtubule-associated inclusions located in the perinuclear region that contain mainly oligomeric, aggregated and ubiquitylated neurotoxic proteins together with ubiquitin-binding protein p62 and chaperones that aid in their formation. Aggresomes are often generated when ubiquitin–proteasome system activity is insufficient. Although they are protective when short-lived, they may be harmful in the long term and can morph into Lewy bodies in Parkinson disease. Aggresomes are cleared by the autophagic–lysosomal network.
- Stress granules
-
Non-membrane enclosed, cytoplasmic agglomerates of ribonucleoproteins that store and protect mRNA during short-term cellular stress. Chaperones such as heat shock 70 kDa protein (HSP70) are involved in assembly and unfolding. In neurodegenerative diseases of ageing, neurotoxic proteins prolong the presence of stress granules and decrease their solubility, leading to aggregation or transformation into aggresomes.
- Peroxisomes
-
Small (100 nm to 1 μm) organelles that oxidize long-chain fatty acids and aid in detoxification. They can be generated by budding-off from the endoplasmic reticulum and replicate via fission. Pexophagy refers to the autophagy of peroxisomes.
- Autophagy-related genes
-
(ATGs). Genes and the molecular machinery for autophagy were characterized in yeast by Y. Ohsumi and others. The associated genes, identified using mutants, were originally termed Apg1–Apg15, yet ATG is now used. In view of conservation across species, this terminology is used for genes and/or proteins that regulate autophagy in humans as well.
- 5′-AMP-activated protein kinase
-
(AMPK). An enzyme involved in energy and nutrient sensing. When activated, AMPK triggers glucose uptake, lipogenesis and triglyceride synthesis. It is a major protein for sensing ATP deficits and initiating the autophagic–lysosomal network.
- Mammalian target of rapamycin complex 1
-
(mTORC1). A multi-tasking serine/threonine-protein kinase that inhibits autophagy, mitophagy and proteasomal degradation. It also has other roles in, for example, controlling mRNA translation and protein synthesis. Mammalian target of rapamycin (mTOR) is part of mTORC1 together with several other regulatory and effector proteins.
- Nicotinamide adenine dinucleotide
-
(NAD) A dinucleotide co-enzyme necessary for energy generation in all cell types. It is a cofactor for activation of NAD-dependent protein deacetylase sirtuin 1 (SIRT1) and is required for operation of the autophagic–lysosomal network. The oxidized and active form is NAD+.
- Acetyl CoA
-
A cofactor involved in protein, carbohydrate and lipid metabolism. It is formed during glycolysis. It provides the acetyl group used by acetyltransferases like histone acetyltransferase p300 to acetylate autophagy-related gene (ATG) proteins, histones and other substrates such as tau.
- RAS-related protein RAB7
-
(RAB7). A member of the GTPase RAS superfamily of monomeric G proteins, which participate in vesicular trafficking, vesicle formation, vesicle movement (actin-mediated and/or tubulin-mediated) and vesicular fusion, as in autophagosomal fusion with lysosomes.
- SNARE
-
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) refers to a complex of proteins including synaptobrevin, syntaxin, synaptosomal-associated protein 25 (SNAP-25) and synaptogamin. SNARE contributes to vesicle fusion by 'zippering' a donor vesicle (such as an autophagosome) onto the recipient compartment (such as the lysosome).
- Phospholipase D1
-
(PLD1). Enzyme involved in the transformation of various lipids; it participates in the fusion of autophagosomes with lysosomes.
- Lysosomal storage disorders
-
(LSDs). Diseases resulting from genetic mutations that lead to failure of lysosomal digestion and consequent accumulation of lipids, proteins and other non-digested material. Their pathology is not restricted to the brain, and the age of onset is much earlier than for sporadic, age-related neurodegenerative disorders.
- Niemann–Pick type C disease
-
A lysosomal storage disorder triggered by a defect in the NPC1 gene responsible for cholesterol transport. Patients often display amyloid-β fragment 42 (Aβ42) and tau pathology, underpinning parallels to Alzheimer disease, in which cholesterol transport is likewise disrupted.
- Heat shock cognate 71 kDa protein
-
(HSC70). A constitutively expressed chaperone that affects ATP-dependent nascent and/or unfolded protein folding. It specifically recognizes proteins with an exposed KFERQ-like sequence and delivers them to lysosome-associated membrane glycoprotein 2A (LAMP2A) on lysosomes, where, aided by other proteins, substrates are translocated to the lumen for degradation by chaperone-mediated autophagy.
- KFERQ
-
The KFERQ motif on a protein is the principal criterion for capture followed by chaperone-mediated autophagy (CMA). Q refers to glutamine, although this sometimes may be an asparagine (N). The other residues are acidic (D and E), basic (K and R) or basic and/or hydrophobic (F). There are, however, variations, and post-translational modification can modify susceptibility of proteins bearing a KFERQ signal for CMA.
- Amyloid precursor protein
-
(APP). A transmembrane protein highly expressed in neurons and involved in maintaining cell–cell contact. Successive cleavage by β-secretases and γ-secretases results in the formation of APP terminal fragments like C99, as well as amyloid-β fragment 42 (Aβ42) and related species of neurotoxic peptide.
- Lipofuscin
-
A pigmented cellular inclusion composed of undigestedlysosomal contents, including oxidized and crosslinked proteins. This electron-dense, autofluorescent material is characteristic of ageing and neurodegenerative disorders of ageing and can be seen in all types of cerebral cell.
- Unfolded protein response
-
(UPR). A protective response to help cells recover from cellular and endoplasmic reticulum stress. The UPR acts via three key effector proteins to modify gene transcription/mRNA translation. The UPR interrupts bulk protein synthesis, promotes the generation of chaperones for protein folding and increases degradation of misfolded proteins. Overactivation and protracted engagement of the UPR are harmful for neurons and are implicated in neurodegenerative disorders of ageing.
- ALN dysfunction
-
Underactive autophagy is a term used when rates of autophagosome formation and cargo sequestration decrease below basal levels or fail to upregulate sufficiently under stress. Impaired autophagy occurs when lysosomal delivery, fusion or digestion of autophagosomes are compromised. Overactive autophagy is the overproduction of autophagosomes and excess autophagic–lysosomal network activity; this can lead to autosis.
- Autosis
-
Autophagy-related cell death mediated principally by the Na+/K+-ATPase pump. This can occur with prolonged and excessive autophagy. It is triggered by hypoxia–ischaemia (as in stroke or traumatic brain injury), but its occurrence in neurodegenerative disorders of ageing is uncertain.
- Apolipoprotein E allele 4
-
(APOE4). A robust genetic risk factor for Alzheimer disease compared with the more common APOE2 and APOE3 alleles. APOE is mainly secreted by astrocytes and binds lipids such as cholesterol, which are carried to neurons. APOE4 is also involved in transport of cholesterol-bound amyloid-β (Aβ) to the blood–brain barrier (APOE4 is less efficient than APOE2 or APOE3) and in driving synthesis of Aβ fragment 42 (Aβ42) (APOE4 is more potent than APOE2 or APOE3).
- Presenilin 1
-
(PS1). A gene encoding the catalytic unit of the γ-secretase complex, which processes amyloid precursor protein (APP) into amyloid-β (Aβ). Mutations are associated with familial Alzheimer disease and in part reflect altered APP processing. In addition, reduced lysosomal acidification and autophagic–lysosomal network function may be involved owing to mutant presenilin 1-driven deficits in maturation and translocation of vacuolar-type H+-ATPase complex (v-ATPase) subunits to the lysosome.
- Parkin
-
A component of the E3 ubiquitin ligase complex that binds to its partner PTEN-induced putative kinase protein 1 (PINK1) to facilitate the autophagic removal of dysfunctional mitochondria that have lost their membrane potential.
- Gaucher disease
-
A primary, autosomal recessive lysosomal storage disease caused by mutations in the GBA1 gene, which encodes β-glucocerebrosidase (βGCase). There is a fivefold higher risk of Parkinson disease (PD) in affected carriers. The activity of βGCase is impaired in a subpopulation of patients with non-familial PD, which includes many patients who have genetic mutations related to lysosomal disruption.
- Superoxide dismutase 1
-
(SOD1). A mitochondrial enzyme dedicated to the reduction of free radicals (reactive oxygen species). SOD1 mutations and dysfunction are seen in a subset of patients with amyotrophic lateral sclerosis.
- CAG-expansion repeats
-
Proteins can contain multiple CAG repeats (CAG encodes glutamine (symbol Q)). When the number of CAG repeats is above normal (for example, greater than 35 for the huntingtin (Htt) protein), proteins aggregate, provoke cellular damage and trigger inherited, polyglutamine diseases such as Huntington disease, spinocerebellar ataxia 3/Joseph–Machado disease (involving the ataxin 3 protein) and spinal and bulbar muscular atrophy (involving the androgen receptor).
- TAT–beclin 1
-
A synthetic peptide comprising 11 amino acids of the HIV transactivator of transcription (TAT) protein transduction domain, a diglycine linker and a (commonly 11-mer) sequence derived from amino acids 267–284 of beclin 1. It is cell penetrant and triggers autophagic–lysosomal network-mediated neurotoxic protein clearance without causing cytotoxicity, although higher concentrations may carry the risk of autosis.
- Heat shock factor 1
-
(HSF1). A protein that occurs as a monomer in the nucleus and cytoplasm and is repressed by heat shock proteins such as heat shock 70 kDa protein (HSP70). Following disruption of proteostasis, heat shock proteins dissociate from HSF1 in order to aid protein folding. HSF1 then trimerizes and acts as a transcription factor to increase production of HSP70 and other neuroprotective proteins.
- Exosomes
-
Small (30–120 nm), ceramide-rich vesicles formed mainly from multivesicular bodies. They are released with their contents (proteins, lipids, sugars and nucleic acids) into the extracellular space upon fusion with the plasma membrane. Exosomes contribute to the spread of neurotoxic proteins. Exosomes in cerebrospinal fluid, blood and urine are stable and useful as biomarkers.
- Immunotherapies
-
Biological therapies that passively or actively boost the body's natural defences. Specific classes of antibody aim to neutralize neurotoxic proteins such as amyloid-β fragment 42 (Aβ42) or tau. Entrance to the brain is limited, but these antibodies may also act as a peripheral sink for neurotoxic proteins in the circulation. In the brain, antibodies probably act for the most part extrinsically to neurons.
Rights and permissions
About this article
Cite this article
Boland, B., Yu, W., Corti, O. et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat Rev Drug Discov 17, 660–688 (2018). https://doi.org/10.1038/nrd.2018.109
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2018.109
This article is cited by
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Reduction of DHHC5-mediated beclin 1 S-palmitoylation underlies autophagy decline in aging
Nature Structural & Molecular Biology (2024)
-
Altered perivascular spaces in subcortical white matter in Parkinson’s disease patients with levodopa-induced dyskinesia
npj Parkinson's Disease (2024)
-
Proteostasis failure exacerbates neuronal circuit dysfunction and sleep impairments in Alzheimer’s disease
Molecular Neurodegeneration (2023)
-
Defective lysosomal acidification: a new prognostic marker and therapeutic target for neurodegenerative diseases
Translational Neurodegeneration (2023)