Key Points
-
Recently generated germline transgenic and knockout mice have provided a detailed view of the implication of the gain or loss of individual microRNAs (miRNAs), miRNA clusters and the miRNA processing machinery in cancer. These have been classified as oncogenic (such as miR-155, miR-21 and miR-17∼92), tumour-suppressive (such as miR-15∼16, LIN28, DICER) or context-dependent (such as miR-146 and miR-29).
-
miRNAs and the miRNA processing machinery are involved in all stages of metastatic disease. Some — such as miR-200, the LIN28–let-7 interaction and DICER — contribute to both metastasis and primary tumour development, whereas others, such as miR-31 and miR-10b, are unique to metastasis.
-
Studies uncovering miRNA function have led to their therapeutic application. Delivering tumour-suppressive miRNAs and silencing oncogenic miRNAs with antagomirs has been successful in various mouse models. Many of these studies began with overexpression and knockdown strategies and have since progressed to delivering miRNA-based molecules intranasally, intratumorally or systemically.
-
Expanding on their therapeutic application, miRNAs are also being evaluated for their ability to sensitize cancers to chemotherapeutics. Much of this work is being accomplished in cell culture, with the hope that it will soon transition into preclinical model systems.
Abstract
In normal cells multiple microRNAs (miRNAs) converge to maintain a proper balance of various processes, including proliferation, differentiation and cell death. miRNA dysregulation can have profound cellular consequences, especially because individual miRNAs can bind to and regulate multiple mRNAs. In cancer, the loss of tumour-suppressive miRNAs enhances the expression of target oncogenes, whereas increased expression of oncogenic miRNAs (known as oncomirs) can repress target tumour suppressor genes. This realization has resulted in a quest to understand the pathways that are regulated by these miRNAs using in vivo model systems, and to comprehend the feasibility of targeting oncogenic miRNAs and restoring tumour-suppressive miRNAs for cancer therapy. Here we discuss progress in using mouse models to understand the roles of miRNAs in cancer and the potential for manipulating miRNAs for cancer therapy as these molecules make their way towards clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chalfie, M., Horvitz, H. R. & Sulston, J. E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24, 59–69 (1981).
Hodgkin, J., Horvitz, H. R. & Brenner, S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91, 67–94 (1979).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Reinhart, B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000).
Slack, F. J. et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669 (2000).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011).
Calin, G. A. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA 101, 2999–3004 (2004).
Ryan, B. M., Robles, A. I. & Harris, C. C. Genetic variation in microRNA networks: the implications for cancer research. Nature Rev. Cancer 10, 389–402 (2010).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nature Rev. Cancer 6, 857–866 (2006).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs — microRNAs with a role in cancer. Nature Rev. Cancer 6, 259–269 (2006).
Costinean, S. et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl Acad. Sci. USA 103, 7024–7029 (2006).
O'Connell, R. M. et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619 (2010).
Rodriguez, A. et al. Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611 (2007).
Thai, T. H. et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608 (2007).
Tili, E. et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc. Natl Acad. Sci. USA 108, 4908–4913 (2011).
Chan, J. A., Krichevsky, A. M. & Kosik, K. S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 (2005).
Landgraf, P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA 103, 2257–2261 (2006).
Hatley, M. E. et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 18, 282–293 (2010).
Ma, X. et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl Acad. Sci. USA 108, 10144–10149 (2011).
Medina, P. P., Nolde, M. & Slack, F. J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86–90 (2010). In addition to references 19 and 20, this study generated miR-21 transgenic mice. All three studies report miR-21 as an oncogene in different contexts.
Wang, J. et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila) 2, 807–813 (2009).
Ventura, A. et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 (2008).
Xiao, C. et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature Immunol. 9, 405–414 (2008).
Castellano, L. et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl Acad. Sci. USA 106, 15732–15737 (2009).
Hayashita, Y. et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65, 9628–9632 (2005).
He, L. et al. A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 (2005).
Lanza, G. et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer 6, 54 (2007).
Mestdagh, P. et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene 29, 1394–1404 (2010).
O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005).
Poliseno, L. et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal 3, ra29 (2010).
Mu, P. et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23, 2806–2811 (2009).
Olive, V. et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 23, 2839–2849 (2009). References 32 and 33 isolated the oncogenic role of miR-19 from the miR-17∼92 cluster. Both studies confirmed that miR-19 was sufficient to promote MYC-induced lymphomagenesis.
Mavrakis, K. J. et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nature Genet. 43, 673–678 (2011).
Calin, G. A. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 99, 15524–15529 (2002).
Raveche, E. S. et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood 109, 5079–5086 (2007).
Salerno, E. et al. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol. Cancer Ther. 8, 2684–2692 (2009).
Klein, U. et al. The DLEU2/miR-15a/16–11 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40 (2010). In this study, the tumour-suppressive contribution of miR-15a∼16 was teased apart from its host gene, Dleu2 . Overexpression of miR15a∼16 could reduce cellular proliferation in Dleu2; mir-15a ∼ 16 double knockout mice.
Musumeci, M. et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 30, 4231–4242 (2011).
Roccaro, A. M. et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 113, 6669–6680 (2009).
Acevedo, V. D., Ittmann, M. & Spencer, D. M. Paths of FGFR-driven tumorigenesis. Cell Cycle 8, 580–588 (2009).
Dahiya, N. et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE 3, e2436 (2008).
He, H. et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA 102, 19075–19080 (2005).
Wang, X. et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 3, e2557 (2008).
Boldin, M. P. et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 (2011).
Starczynowski, D. T. et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp. Hematol. 39, 167–178 (2011).
Zhao, J. L. et al. NF- κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl Acad. Sci. USA 108, 9184–9189 (2011). A careful analysis of mir-146a -knockout mice. Mice developed myeloid sarcomas and lymphomas. A link between miR-146a and NF-κB was also described.
Garcia, A. I. et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 3, 279–290 (2011).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-kB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Dickson, K. M., Bhakar, A. L. & Barker, P. A. TRAF6-dependent NF-kB transcriptional activity during mouse development. Dev. Dyn. 231, 122–127 (2004).
Thomas, J. A. et al. Impaired cytokine signaling in mice lacking the IL-1 receptor-associated kinase. J. Immunol. 163, 978–984 (1999).
Fabbri, M. et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA 104, 15805–15810 (2007).
Mott, J. L., Kobayashi, S., Bronk, S. F. & Gores, G. J. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26, 6133–6140 (2007).
Pekarsky, Y. et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 66, 11590–11593 (2006).
Zhao, J. J. et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 115, 2630–2639 (2010).
Garzon, R. et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 114, 5331–5341 (2009).
Pekarsky, Y. & Croce, C. M. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 1, 224–227 (2010).
Han, Y. C. et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 207, 475–489 (2010).
Santanam, U. et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc. Natl Acad. Sci. USA 107, 12210–12215 (2010).
Zhu, H. et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature Genet. 42, 626–630 (2010).
Zhu, H. et al. The Lin28/let-7 Axis Regulates Glucose Metabolism. Cell 147, 81–94 (2011).
Trang, P. et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 29, 1580–1587 (2009). This was the first indication that let-7 could be used to reduce preformed lung tumours in mice, in support of its clinical application.
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Kumar, M. S. et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23, 2700–2704 (2009).
Lambertz, I. et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 17, 633–641 (2010).
Martello, G. et al. A MicroRNA targeting dicer for metastasis control. Cell 141, 1195–1207 (2010). This paper describes a direct link between high miR-103 and miR-107 expression and low DICER expression in cancer. miR-103 and miR-107 abrogate DICER expression,leading to reduced miRNA biogenesis and acceleratedmetastasis.
Su, X. et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 (2010). This paper reports that TAp63 suppresses tumorigenesis and metastasis by transactivating DICER expression.
Nicoloso, M. S., Spizzo, R., Shimizu, M., Rossi, S. & Calin, G. A. MicroRNAs — the micro steering wheel of tumour metastases. Nature Rev. Cancer 9, 293–302 (2009).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008).
Park, S. M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Gibbons, D. L. et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 23, 2140–2151 (2009).
Kong, D. et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS ONE 5, e12445 (2010).
Wellner, U. et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495 (2009).
Korpal, M., Lee, E. S., Hu, G. & Kang, Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008).
Viswanathan, S. R., Daley, G. Q. & Gregory, R. I. Selective blockade of microRNA processing by Lin28. Science 320, 97–100 (2008).
Viswanathan, S. R. et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genet. 41, 843–848 (2009).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Dangi-Garimella, S. et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 28, 347–358 (2009).
Li, X. et al. MicroRNA-107, an oncogene microRNA that regulates tumor invasion and metastasis by targeting DICER1 in gastric cancer: miR-107 promotes gastric cancer invasion and metastasis. J. Cell. Mol. Med. 15, 1887–1895 (2011).
Tokumaru, S., Suzuki, M., Yamada, H., Nagino, M. & Takahashi, T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29, 2073–2077 (2008).
Valastyan, S., Chang, A., Benaich, N., Reinhardt, F. & Weinberg, R. A. Concurrent suppression of integrin α5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 70, 5147–5154 (2010).
Valastyan, S., Chang, A., Benaich, N., Reinhardt, F. & Weinberg, R. A. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev. 25, 646–659 (2011). A comprehensive study evaluating the antimetastatic role of miR-31 and suggesting the potential use of miR-31 for the treatment of metastatic disease.
Valastyan, S. et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137, 1032–1046 (2009).
Ma, L. et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotech. 28, 341–347 (2010). The only report of systemically delivering antagomirs for cancer therapy. In mice, anti-miR-10b treatment suppressed metastasis without affecting primary tumour development.
Huang, Q. et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 10, 202–210 (2008).
Tavazoie, S. F. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008).
Roush, S. & Slack, F. J. The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516 (2008).
Dangi-Garimella, S., Strouch, M. J., Grippo, P. J., Bentrem, D. J. & Munshi, H. G. Collagen regulation of let-7 in pancreatic cancer involves TGF-β1-mediated membrane type 1-matrix metalloproteinase expression. Oncogene 30, 1002–1008 (2011).
Wang, M., Hu, Y., Amatangelo, M. D. & Stearns, M. E. Role of ribosomal protein RPS2 in controlling let-7a expression in human prostate cancer. Mol. Cancer Res. 9, 36–50 (2011).
Trang, P. et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19, 1116–1122 (2011). This paper reports the use of systemically delivered miRNAs ( let-7 and miR-34) to treat preformed lung tumours. Systemically delivered miRNAs were well tolerated and effective.
Lee, S. T. et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J. Neurooncol. 102, 19–24 (2010).
Yu, F. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 (2007).
Hirai, H. et al. Activation of the c-K-ras oncogene in a human pancreas carcinoma. Biochem. Biophys. Res. Commun. 127, 168–174 (1985).
Kent, O. A. et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 24, 2754–2759 (2010).
Nakano, H. et al. Isolation of transforming sequences of two human lung carcinomas: structural and functional analysis of the activated c-K-ras oncogenes. Proc. Natl Acad. Sci. USA 81, 71–75 (1984).
Pramanik, D. et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 10, 1470–1480 (2011). This study reports the systemic delivery of miR-34, miR-143 and miR-145 to orthotopic pancreatic tumours in vivo . Tumour growth was inhibited with no signs of toxicity. This is especially interesting because blood flow to the pancreas is thought to be low.
Earle, J. S. et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J. Mol. Diagn. 12, 433–440 (2010).
Gao, W. et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed. Pharmacother. 64, 399–408 (2010).
Han, Y. et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS ONE 6, e18286 (2011).
Zhang, X., Liu, S., Hu, T., He, Y. & Sun, S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50, 490–499 (2009).
Marchini, S. et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 12, 273–285 (2011).
Volinia, S. et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 20, 589–599 (2010).
Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199 (2009).
Kasinski, A. L. & Slack, F. J. Potential microRNA therapies targeting Ras, NFκB and p53 signaling. Curr. Opin. Mol. Ther. 12, 147–157 (2010).
Brosh, R. & Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nature Rev. Cancer 9, 701–713 (2009).
Wiggins, J. F. et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930 (2010).
Liu, C. et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Med. 17, 211–215 (2011). This study showed that systemic miR-34 delivery inhibited prostate cancer metastasis and increased survival. CD44 was validated as a causal target of the miR-34 therapeutic effect.
Komar, G. et al. Decreased blood flow with increased metabolic activity: a novel sign of pancreatic tumor aggressiveness. Clin. Cancer Res. 15, 5511–5517 (2009).
Bai, S. et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 284, 32015–32027 (2009).
Burchard, J. et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol. Syst. Biol. 6, 402 (2010).
Tsai, W. C. et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49, 1571–1582 (2009).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Su, J., Baigude, H., McCarroll, J. & Rana, T. M. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 39, e38 (2011).
Kota, J. et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017 (2009).
Ji, J. et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361, 1437–1447 (2009).
Lu, J. et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 71, 225–233 (2011).
Sander, S., Bullinger, L. & Wirth, T. Repressing the repressor: a new mode of MYC action in lymphomagenesis. Cell Cycle 8, 556–559 (2009).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005).
Jeon, H. M. et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 71, 3410–3421 (2011).
Cittelly, D. M. et al. Oncogenic HER2Δ16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis 31, 2049–2057 (2010).
Cittelly, D. M. et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 9, 317 (2010).
Miller, T. E. et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 283, 29897–29903 (2008).
Zhong, M., Ma, X., Sun, C. & Chen, L. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem. Biol. Interact. 184, 431–438 (2010).
Harris, T. J. & McCormick, F. The molecular pathology of cancer. Nature Rev. Clin. Oncol. 7, 251–265 (2010).
Kastl, L., Brown, I. & Schofield, A. C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 12 Mar 2011 (doi:10.1007/s10549-011-1424-3).
Johnson, S. M. et al. RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005).
Weidhaas, J. B. et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 67, 11111–11116 (2007).
Boyerinas, B. et al. Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of MDR1. Int. J. Cancer 26 May 2011 (doi:10.1002/ijc.26190).
Chen, F. et al. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 23, 1457–1462 (2010).
Fujita, Y. et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 285, 19076–19084 (2010).
Kojima, K., Fujita, Y., Nozawa, Y., Deguchi, T. & Ito, M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate 70, 1501–1512 (2010).
Zhou, M. et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507 (2010).
Holleman, A. et al. miR-135a contributes to paclitaxel resistance in tumor cells both in vitro and in vivo. Oncogene 30, 4386–4398 (2011). This study identified upregulation of miR-135a in paclitaxel-resistant tumours. Antagonizing miR-135a in vivo resensitized paclitaxel resistant tumours to this therapy.
Schetter, A. J. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299, 425–436 (2008).
Valeri, N. et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl Acad. Sci. USA 107, 21098–21103 (2010).
Tomimaru, Y. et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br. J. Cancer 103, 1617–1626 (2010).
Hwang, J. H. et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE 5, e10630 (2010).
Li, Y. et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 69, 6704–6712 (2009).
Park, J. K., Lee, E. J., Esau, C. & Schmittgen, T. D. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 38, e190–e199 (2009).
Ceppi, P. et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 8, 1207–1216 (2010).
Galluzzi, L. et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 70, 1793–1803 (2010).
Weeraratne, S. D. et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 13, 165–175 (2011).
Ambros, V. microRNAs: tiny regulators with great potential. Cell 107, 823–826 (2001).
Pecot, C. V., Calin, G. A., Coleman, R. L., Lopez-Berestein, G. & Sood, A. K. RNA interference in the clinic: challenges and future directions. Nature Rev. Cancer 11, 59–67 (2011).
Esquela-Kerscher, A. et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7, 759–764 (2008).
Kumar, M. S. et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl Acad. Sci. USA 105, 3903–3908 (2008).
Duncan, R. The dawning era of polymer therapeutics. Nature Rev. Drug Discov. 2, 347–360 (2003).
Estevez, M. C. et al. Nanoparticle-aptamer conjugates for cancer cell targeting and detection. Methods Mol. Biol. 624, 235–248 (2010).
Chen, Y., Zhu, X., Zhang, X., Liu, B. & Huang, L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 18, 1650–1656 (2010).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011).
Hunter, M. P. et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3, e3694 (2008).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Grimm, D. et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006).
Grimm, D. et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 120, 3106–3119 (2010).
Chen, K. & Rajewsky, N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genet. 38, 1452–1456 (2006).
Chin, L. J. et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 68, 8535–8540 (2008).
Jazdzewski, K. et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA 105, 7269–7274 (2008).
Hoffman, A. E. et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 69, 5970–5977 (2009).
Kumar, M. S., Lu, J., Mercer, K. L., Golub, T. R. & Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet. 39, 673–677 (2007).
Acknowledgements
A.L.K. was supported by a US National Institutes of Health (NIH) grant (1F32CA153885-01) and an American Cancer Society Postdoctoral Fellowship (120,766-PF-11-244-01-TBG). F.J.S. was supported by a grant from the US National Cancer Institute (NCI) (1R01CA131301).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
F.J.S. is a consultant to Mirna Therapeutics, Inc. and Mira Dx, Inc. A.L.K. declares no competing financial interests.
Related links
DATABASES
FURTHER INFORMATION
Glossary
- miR-17∼92
-
This is a cluster of miRNAs containing miR-17, miR-20a, miR-18a, miR-19a, miR-19b and miR-92a. Clusters are expressed as a polycistronic message from a single promoter; this is indicated in the text by a tilde (∼).
- Paralogues
-
Genes that are derived from the same ancestral gene but that reside in different locations in the same genome.
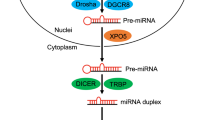
- DICER
-
An endoribonuclease that cleaves precursor microRNAs (pre-miRNAs) into 20–25 nucleotide double-stranded RNAs.
- Adaptive immune response
-
The adaptive immune response — also known as specific or acquired immunity — is mediated by antigen-specific lymphocytes and antibodies.
- Innate immune response
-
The innate immune system provides an immediate, non-specific defence against pathogens until an adaptive, pathogen-specific immune response is able to develop.
- Nestin promoter
-
This promoter drives the expression of genes in the central and peripheral nervous system and in myogenic and other tissues.
- DMBA–TPA model
-
A two-stage chemical skin carcinogenesis model using a single dose of the genotoxic carcinogen, 7,12-dimethylbenz(a) anthracene (DMBA), followed by multiple doses of a non-genotoxic tumour-promoter, 12-O-tetradecanoylphorbol-13-acetate (TPA).
- Papillomas
-
Benign tumours of epithelial origin that grow outwards.
- Polycistronic
-
A single transcript that carries the information of several genes.
- Fragile site
-
A site in a chromosome that is susceptible to chromosome breakage and fusion with other chromosomes.
- Host gene
-
A gene containing another gene (such as a microRNA within an intron of a protein-coding gene).
- Seed region
-
Nucleotides 2–7 of the microRNA, typically having 100% complementarity with the target gene.
- Xenografts
-
Grafts of cells or tissues that are transplanted from one species to another.
- Haploinsufficient
-
A phenotype that arises in diploid organisms owing to the loss of only one allele.
- Hemizygous
-
A state of having a single copy of a gene (in a diploid organism).
- Epithelial to mesenchymal transition
-
(EMT). The conversion of polarized, immotile epithelial cells to motile mesenchymal cells. It is characterized by the loss of adhesion, the repression of E-cadherin, the expression of mesenchymal markers and increased cell motility.
- Adjuvant
-
Treatment given in addition to the primary therapy. For cancer treatment, this typically refers to the therapy given after the surgical resection of a tumour.
- Antagomirs
-
Chemically engineered, cholesterol-conjugated single-stranded RNA oligonucleotides that are complementary to microRNAs.
- Autochthonous
-
Formed from endogenous tissue in the correct anatomical location.
- Orthotopic
-
Transplanted into the correct anatomical location.
- Silent information regulator 1
-
(SIRT1). A class-III histone deacetylase that regulates apoptosis in response to genotoxic and oxidative stress.
- star strand
-
(miRNA*). The passenger strand of the mature microRNA (miRNA), originally thought not to be involved in miRNA-induced silencing; however, deep sequencing followed by functional studies have determined that the some of the miRNA* products are abundant and functional.
- Selective oestrogen receptor modulator
-
(SERM). An agent that targets the oestrogen receptors, ERα and ERβ, which are often upregulated in breast cancer.
- Docetaxel
-
A cytotoxic antimicrotubule agent that is used to treat breast, ovarian, prostate and non-small-cell lung cancer.
- Gemcitabine
-
A nucleoside analogue that is used to treat multiple forms of cancer.
- Cisplatin
-
A platinum-containing cytotoxic cancer drug.
Rights and permissions
About this article
Cite this article
Kasinski, A., Slack, F. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 11, 849–864 (2011). https://doi.org/10.1038/nrc3166
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3166
This article is cited by
-
MicroRNA-188-5p inhibits hepatocellular carcinoma proliferation and migration by targeting forkhead box N2
BMC Cancer (2023)
-
MicroRNA 29a therapy for CEACAM6-expressing lung adenocarcinoma
BMC Cancer (2023)
-
A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy
Oncogene (2023)
-
Role of non-coding RNAs in neuroblastoma
Cancer Gene Therapy (2023)
-
MicroRNA-3692-3p is overexpressed in lung tumors but may not serve as a prognostic biomarker in lung cancer patients
Molecular Biology Reports (2023)