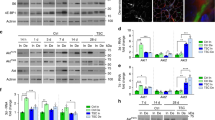
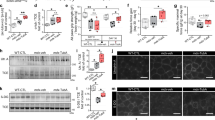
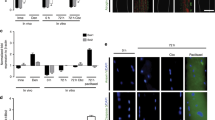
Abstract
Electrical activity arising from motor innervation influences skeletal muscle physiology by controlling the expression of many muscle genes, including those encoding acetylcholine receptor (AChR) subunits. How electrical activity is converted into a transcriptional response remains largely unknown. We show that motor innervation controls chromatin acetylation in skeletal muscle and that histone deacetylase 9 (HDAC9) is a signal-responsive transcriptional repressor which is downregulated upon denervation, with consequent upregulation of chromatin acetylation and AChR expression. Forced expression of Hdac9 in denervated muscle prevents upregulation of activity-dependent genes and chromatin acetylation by linking myocyte enhancer factor 2 (MEF2) and class I HDACs. By contrast, Hdac9-null mice are supersensitive to denervation-induced changes in gene expression and show chromatin hyperacetylation and delayed perinatal downregulation of myogenin, an activator of AChR genes. These findings show a molecular mechanism to account for the control of chromatin acetylation by presynaptic neurons and the activity-dependent regulation of skeletal muscle genes by motor innervation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Guan, Z. et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 111, 483–493 (2002).
Alarcon, J.M. et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42, 947–959 (2004).
Korzus, E., Rosenfeld, M.G. & Mayford, M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972 (2004).
Sanes, J.R. & Lichtman, J.W. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2, 791–805 (2001).
Schaeffer, L., Duclert, N., Huchet-Dymanus, M. & Changeux, J.P. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 17, 3078–3090 (1998).
Fromm, L. & Burden, S.J. Synapse-specific and neuregulin-induced transcription require an ets site that binds GABPα/GABPβ. Genes Dev. 12, 3074–3083 (1998).
Schaeffer, L. de Kerchove d'Exaerde, A. & Changeux, J.P. Targeting transcription to the neuromuscular synapse. Neuron 31, 15–22 (2001).
Gundersen, K., Rabben, I., Klocke, B.J. & Merlie, J.P. Overexpression of myogenin in muscles of transgenic mice: interaction with Id-1, negative crossregulation of myogenic factors, and induction of extrasynaptic acetylcholine receptor expression. Mol. Cell. Biol. 15, 7127–7134 (1995).
Piette, J., Bessereau, J.L., Huchet, M. & Changeux, J.P. Two adjacent MyoD1-binding sites regulate expression of the acetylcholine receptor alpha-subunit gene. Nature 345, 353–355 (1990).
Duclert, A., Piette, J. & Changeux, J.P. Influence of innervation of myogenic factors and acetylcholine receptor alpha-subunit mRNAs. Neuroreport 2, 25–28 (1991).
Witzemann, V. & Sakmann, B. Differential regulation of MyoD and myogenin mRNA levels by nerve induced muscle activity. FEBS Lett. 282, 259–264 (1991).
Eftimie, R., Brenner, H.R. & Buonanno, A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc. Natl. Acad. Sci. USA 88, 1349–1353 (1991).
Merlie, J.P., Mudd, J., Cheng, T.C. & Olson, E.N. Myogenin and acetylcholine receptor alpha gene promoters mediate transcriptional regulation in response to motor innervation. J. Biol. Chem. 269, 2461–2467 (1994).
Bessereau, J.L., Laudenbach, V., Le Poupon, C. & Changeux, J.P. Nonmyogenic factors bind nicotinic acetylcholine receptor promoter elements required for response to denervation. J. Biol. Chem. 273, 12786–12793 (1998).
Sartorelli, V., Huang, J., Hamamori, Y. & Kedes, L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17, 1010–1026 (1997).
Puri, P.L. et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1, 35–45 (1997).
Polesskaya, A. et al. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21, 5312–5320 (2001).
McKinsey, T.A., Zhang, C.L. & Olson, E.N. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11, 497–504 (2001).
Lu, J., McKinsey, T.A., Zhang, C.L. & Olson, E.N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6, 233–244 (2000).
McKinsey, T.A., Zhang, C.L., Lu, J. & Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 (2000).
McKinsey, T.A., Zhang, C.L. & Olson, E.N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97, 14400–14405 (2000).
Dressel, U. et al. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276, 17007–17013 (2001).
McKinsey, T.A., Zhang, C.L. & Olson, E.N. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312–6321 (2001).
Zhou, X., Richon, V.M., Rifkind, R.A. & Marks, P.A. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. USA 97, 1056–1061 (2000).
Zhou, X., Marks, P.A., Rifkind, R.A. & Richon, V.M. Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. USA 98, 10572–10577 (2001).
Sparrow, D.B. et al. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 18, 5085–5098 (1999).
Klarsfeld, A. et al. Regulation of muscle AChR alpha subunit gene expression by electrical activity: involvement of protein kinase C and Ca2+. Neuron 2, 1229–1236 (1989).
Bessereau, J.L., Stratford-Perricaudet, L.D., Piette, J., Le Poupon, C. & Changeux, J.P. In vivo and in vitro analysis of electrical activity-dependent expression of muscle acetylcholine receptor genes using adenovirus. Proc. Natl. Acad. Sci. USA 91, 1304–1308 (1994).
Walke, W., Xiao, G. & Goldman, D. Identification and characterization of a 47 base pair activity-dependent enhancer of the rat nicotinic acetylcholine receptor delta-subunit promoter. J. Neurosci. 16, 3641–3651 (1996).
Lemercier, C. et al. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275, 15594–15599 (2000).
Zhang, C.L., McKinsey, T.A. & Olson, E.N. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc. Natl. Acad. Sci. USA 98, 7354–7359 (2001).
Zhang, C.L. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 (2002).
Zhang, C.L., McKinsey, T.A., Lu, J.R. & Olson, E.N. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276, 35–39 (2001).
Cheng, T.C., Wallace, M.C., Merlie, J.P. & Olson, E.N. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261, 215–218 (1993).
Molkentin, J.D., Black, B.L., Martin, J.F. & Olson, E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83, 1125–1136 (1995).
Black, B.L. & Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196 (1998).
Wolff, J.A. et al. Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468 (1990).
Ornatsky, O.I., Andreucci, J.J. & McDermott, J.C. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272, 33271–33278 (1997).
Naya, F.J., Wu, C., Richardson, J.A., Overbeek, P. & Olson, E.N. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 126, 2045–2052 (1999).
Wu, H. et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 19, 1963–1973 (2000).
Hyatt, J.P., Roy, R.R., Baldwin, K.M. & Edgerton, V.R. Nerve activity-independent regulation of skeletal muscle atrophy: role of MyoD and myogenin in satellite cells and myonuclei. Am. J. Physiol. Cell Physiol. 285, C1161–C1173 (2003).
Tang, J., Jo, S.A. & Burden, S.J. Separate pathways for synapse-specific and electrical activity-dependent gene expression in skeletal muscle. Development 120, 1799–1804 (1994).
Hediger, F. & Burden, S.J. Nuclear organization and silencing: putting things in their place. Nat. Cell Biol. 4, E53–E55 (2002).
Fisher, A.G. & Merkenschlager, M. Gene silencing, cell fate and nuclear organization. Curr. Opin. Genet. Dev. 12, 193–197 (2002).
Baxter, J., Merkenschlager, M. & Fisher, A.G. Nuclear organization and gene expression. Curr. Opin. Cell Biol. 14, 372–376 (2002).
Lemercier, C. et al. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 277, 22045–22052 (2002).
Zhang, C.L., McKinsey, T.A. & Olson, E.N. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22, 7302–7312 (2002).
Xu, Q. et al. p38 Mitogen-activated protein kinase-, calcium-calmodulin-dependent protein kinase-, and calcineurin-mediated signaling pathways transcriptionally regulate myogenin expression. Mol. Biol. Cell 13, 1940–1952 (2002).
McKinsey, T.A., Zhang, C.L. & Olson, E.N. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40–47 (2002).
Chang, S. et al. Histone deacetylase 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24, 8467–8476 (2004).
Acknowledgements
We thank E. Chopin for technical assistance in chromatin immunoprecipitation (ChIP) assay; C. Antos for kind and helpful assistance; and A. Ravel-Chapuis, M. Vandromme, K. Ancelin, A. de Kerchove d'Exaerde, Y.-G. Gangloff and A. Sergeant for fruitful discussions and critical reading of the manuscript. This work was supported by the Association Française contre les Myopathies, the Centre National de la Recherche Scientifique, the Ministere de l'Education Nationale, de la Recherche et de la Technologie, and grants from the U.S. National Institutes of Health, the Texas Advanced Technology Program, The Robert A. Welch Foundation and the Donald W. Reynolds Foundation to E.N.O. and R.B.D.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
HDAC 4, 5 and MITR localization in skeletal muscle. (PDF 215 kb)
Supplementary Fig. 2
Myogenin and AChR α subunit are expressed at the same levels in wild-type and Hdac9−/− mice. (PDF 101 kb)
Rights and permissions
About this article
Cite this article
Méjat, A., Ramond, F., Bassel-Duby, R. et al. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci 8, 313–321 (2005). https://doi.org/10.1038/nn1408
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn1408
This article is cited by
-
Epigenetic regulation of beta-endorphin synthesis in hypothalamic arcuate nucleus neurons modulates neuropathic pain in a rodent pain model
Nature Communications (2023)
-
Motoneurons innervation determines the distinct gene expressions in multinucleated myofibers
Cell & Bioscience (2022)
-
Common protein-coding variants influence the racing phenotype in galloping racehorse breeds
Communications Biology (2022)
-
Histone deacetylase (HDAC) 9: versatile biological functions and emerging roles in human cancer
Cellular Oncology (2021)
-
Signatures of selection in the genome of Swedish warmblood horses selected for sport performance
BMC Genomics (2019)