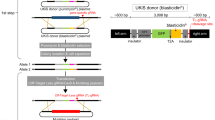
Abstract
Recent consolidation of the whole-genome sequence with genome-wide transcriptome profiling revealed the existence of functional units within the genome in specific chromosomal regions, as seen in the coordinated expression of gene clusters and colocalization of functionally related genes. An efficient region-specific mutagenesis screen would greatly facilitate research in addressing the importance of these clusters. Here we use the 'local hopping' phenomenon of a DNA-type transposon, Sleeping Beauty (SB), for region-specific saturation mutagenesis. A transgenic mouse containing both transposon (acts as a mutagen) and transposase (recognizes and mobilizes the transposon) was bred for germ-cell transposition events, allowing us to generate many mutant mice. All genes within a 4-Mb region of the original donor site were mutated by SB, indicating the potential of this system for functional genomic studies within a specific chromosomal region.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Versteeg, R. et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13, 1998–2004 (2003).
Caron, H. et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291, 1289–1292 (2001).
Kmita, M. & Duboule, D. Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333 (2003).
Palstra, R.J. et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 (2003).
Maclean, J.A., II et al. Rhox: a new homeobox gene cluster. Cell 120, 369–382 (2005).
Russell, W.L. et al. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA 76, 5818–5819 (1979).
Hitotsumachi, S., Carpenter, D.A. & Russell, W.L. Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc. Natl. Acad. Sci. USA 82, 6619–6621 (1985).
Hansen, J. et al. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA 100, 9918–9922 (2003).
Carlson, C.M. & Largaespada, D.A. Insertional mutagenesis in mice: new perspectives and tools. Nat. Rev. Genet. 6, 568–580 (2005).
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98, 9191–9196 (2001).
Horie, K. et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23, 9189–9207 (2003).
Carlson, C.M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Dupuy, A.J., Fritz, S. & Largaespada, D.A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001).
Fischer, S.E., Wienholds, E. & Plasterk, R.H. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98, 6759–6764 (2001).
Luo, G., Ivics, Z., Izsvak, Z. & Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95, 10769–10773 (1998).
Ivics, Z., Hackett, P.B., Plasterk, R.H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997).
Smits, P. et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1, 277–290 (2001).
Yi, S.E., Daluiski, A., Pederson, R., Rosen, V. & Lyons, K.M. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 127, 621–630 (2000).
Yi, S.E. et al. The type I BMP receptor BmprIB is essential for female reproductive function. Proc. Natl. Acad. Sci. USA 98, 7994–7999 (2001).
Karolchik, D. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 31, 51–54 (2003).
Kiyosawa, H. et al. Systematic genome-wide approach to positional candidate cloning for identification of novel human disease genes. Intern. Med. J. 34, 79–90 (2004).
Kile, B.T. et al. Functional genetic analysis of mouse chromosome 11. Nature 425, 81–86 (2003).
Yant, S.R. et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 25, 2085–2094 (2005).
Collier, L.S., Carlson, C.M., Ravimohan, S., Dupuy, A.J. & Largaespada, D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005).
Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G. & Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005).
Ayyanathan, K. et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17, 1855–1869 (2003).
Kokubu, C. et al. Undulated short-tail deletion mutation in the mouse ablates Pax1 and leads to ectopic activation of neighboring Nkx2–2 in domains that normally express Pax1. Genetics 165, 299–307 (2003).
Auwerx, J. et al. The European dimension for the mouse genome mutagenesis program. Nat. Genet. 36, 925–927 (2004).
Austin, C.P. et al. The knockout mouse project. Nat. Genet. 36, 921–924 (2004).
Devon, R.S., Porteous, D.J. & Brookes, A.J. Splinkerettes—improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23, 1644–1645 (1995).
Acknowledgements
We thank G. Kondoh, M. Kouno, E.S. Saito and R. Ikeda for helpful advice and excellent technical assistance. We also thank S. Makino and Y. Odan for administrative assistance in preparing this manuscript. This work was supported by grants from the New Energy and Industrial Technology Development Organization of Japan; Uehara Memorial Foundation; Preventure Program, Japan Science and Technology Agency; RIKEN, The Institute of Physical and Chemical Research; and a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Examples of two phenotypic screens generated by SB transposon system. (PDF 72 kb)
Supplementary Fig. 2
Determining the exact location of IR3 donor site on chromosome 12. (PDF 85 kb)
Supplementary Fig. 3
Gene trap and region-specific saturation mutagenesis using the SB transposon system for IF2 donor mouse. (PDF 214 kb)
Supplementary Fig. 4
Detection of new transcripts using the SB transposon system. (PDF 90 kb)
Supplementary Fig. 5
Screen shot of the UCSC Mouse Genome Browser Version mm4 with our SB database overview of transposon insertion sites for IR3 and IF2 donor mice. (PDF 136 kb)
Supplementary Table 1
List of primers used in the nested-PCR. (PDF 54 kb)
Rights and permissions
About this article
Cite this article
Keng, V., Yae, K., Hayakawa, T. et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods 2, 763–769 (2005). https://doi.org/10.1038/nmeth795
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth795
This article is cited by
-
Insertional mutagenesis using the Sleeping Beauty transposon system identifies drivers of erythroleukemia in mice
Scientific Reports (2019)
-
Chromatin states shape insertion profiles of the piggyBac, Tol2 and Sleeping Beauty transposons and murine leukemia virus
Scientific Reports (2017)
-
Reduced CaM Kinase II and CaM Kinase IV Activities Underlie Cognitive Deficits in NCKX2 Heterozygous Mice
Molecular Neurobiology (2017)
-
Ds transposon is biased towards providing splice donor sites for exonization in transgenic tobacco
Plant Molecular Biology (2012)
-
The mouse genetics toolkit: revealing function and mechanism
Genome Biology (2011)