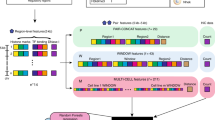
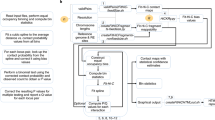
Abstract
Hi-C is a genome-wide sequencing technique used to investigate 3D chromatin conformation inside the nucleus. Computational methods are required to analyze Hi-C data and identify chromatin interactions and topologically associating domains (TADs) from genome-wide contact probability maps. We quantitatively compared the performance of 13 algorithms in their analyses of Hi-C data from six landmark studies and simulations. This comparison revealed differences in the performance of methods for chromatin interaction identification, but more comparable results for TAD detection between algorithms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Cavalli, G. & Misteli, T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 20, 290–299 (2013).
Dixon, J.R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E.P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Rao, S.S.P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Schmitt, A.D., Hu, M. & Ren, B. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol. 17, 743–755 (2016).
Ay, F. & Noble, W.S. Analysis methods for studying the 3D architecture of the genome. Genome Biol. 16, 183 (2015).
Mora, A., Sandve, G.K., Gabrielsen, O.S. & Eskeland, R. In the loop: promoter-enhancer interactions and bioinformatics. Brief. Bioinform. 17, 980–995 (2016).
Shavit, Y., Merelli, I., Milanesi, L. & Lio', P. How computer science can help in understanding the 3D genome architecture. Brief. Bioinform. 17, 733–744 (2016).
Durand, N.C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Ay, F., Bailey, T.L. & Noble, W.S. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 24, 999–1011 (2014).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Hwang, Y.C. et al. HIPPIE: a high-throughput identification pipeline for promoter interacting enhancer elements. Bioinformatics 31, 1290–1292 (2015).
Lun, A.T.L. & Smyth, G.K. diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinformatics 16, 258 (2015).
Lévy-Leduc, C., Delattre, M., Mary-Huard, T. & Robin, S. Two-dimensional segmentation for analyzing Hi-C data. Bioinformatics 30, i386–i392 (2014).
Serra, F., Baù, D., Filion, G. & Marti-Renom, M.A. Structural features of the fly chromatin colors revealed by automatic three-dimensional modeling. Preprint at http://dx.doi.org/10.1101/036764 (2016).
Crane, E. et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244 (2015).
Weinreb, C. & Raphael, B.J. Identification of hierarchical chromatin domains. Bioinformatics 32, 1601–1609 (2016).
Filippova, D., Patro, R., Duggal, G. & Kingsford, C. Identification of alternative topological domains in chromatin. Algorithms Mol. Biol. 9, 14 (2014).
Dixon, J.R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Yaffe, E. & Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Sauria, M.E.G., Phillips-Cremins, J.E., Corces, V.G. & Taylor, J. HiFive: a tool suite for easy and efficient HiC and 5C data analysis. Genome Biol. 16, 237 (2015).
Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Ho, J.W.K. et al. Comparative analysis of metazoan chromatin organization. Nature 512, 449–452 (2014).
Dali, R. & Blanchette, M. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 45, 2994–3005 (2017).
Imakaev, M.V., Fudenberg, G. & Mirny, L.A. Modeling chromosomes: beyond pretty pictures. FEBS Lett. 589, 3031–3036 (2015).
Dekker, J. et al. The 4D nucleome project. Preprint at: http://dx.doi.org/10.1101/103499 (2017).
Schoenfelder, S. et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 25, 582–597 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Acknowledgements
This work was supported by AIRC Special Program Molecular Clinical Oncology “5 per mille” (to S.B.); by AIRC Start-up grant 2015 N.16841 (to F.F.); and by Italian Epigenomics Flagship Project (Epigen) (to S.B.). This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Program (grant agreement no. 670126-DENOVOSTEM to S.B. and M.F.) and from CINECA (ISCRA Class C project HP10CDMGT8 to M.F.). C.M.L. is supported by SIPOD (Structured International Post Doc program of SEMM), a Marie Curie cofunded fellowship. We thank A. Lun (University of Cambridge) for sharing the code used to simulate Hi-C data in the diffHic article. We thank F. Fanelli (Dept. of Life Sciences, University of Modena and R. Emilia) and the center for scientific computing of the University of Modena and R. Emilia for the use of GPUs. We thank M. Cordenonsi (Dept. of Molecular Medicine, University of Padova), P. Maiuri (The FIRC Institute of Molecular Oncology, IFOM), E. Sebestyen (The FIRC Institute of Molecular Oncology, IFOM), and M. Morelli (Center for Genomic Science, Istituto Italiano di Tecnologia IIT) for critical feedback on the manuscript. We would also like to thank the authors of all the tools compared for providing support for their methods and for prompt replies to our inquiries.
Author information
Authors and Affiliations
Contributions
M.F., C.N., and K.P. collected the experimental data and implemented the computational pipelines. M.F., C.N., K.P., and C.M.L. analyzed the Hi-C data sets. M.F. and C.N. compiled the list of interaction evidences. F.F. generated the simulated data. M.F., F.F., and S.B. designed the experiments and analyzed the results. M.F., C.N., F.F., and S.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Number of cis and trans interactions called by each method versus the number of reads.
a) Scatter plot of total number of cis interactions called by each method versus the number of reads retained by the filtering step in all datasets at 1Mb, 40kb, and 5kb resolutions. Different points represent sample replicates. Linear interpolation (of log transformed data) is shown as solid line only for datasets at 5kb, where more data points are available. b) Same as in a) for trans interactions. Fit-Hi-C and HiCCUPS do not return trans interactions. c) Same as in b) for the ratio of cis over trans interactions in datasets at 5kb.
Supplementary Figure 2 Average distance between anchoring points in cis interactions for datasets at 1Mb and 40kb resolution and representative heatmaps.
a) Boxplot of average distances between anchoring points in cis interactions (log scale) in sample replicates of all datasets at 1Mb and 40kb resolutions. At 1Mb (Lieberman-Aiden dataset), HIPPIE found just 1 interaction between two adjacent bins. b) Heatmap of the contact matrix of Lieberman Aiden replicate A_NcoI (chr1:20,000,000-120,000,000) at 1Mb resolution. Identified peaks are marked in different colors for the various methods. HIPPIE recalled no interactions in this region. c) Heatmaps of the contact matrix of Dixon 2012 H1-hESC replicate B (chr21:30,000,000-40,000,000) at 40kb resolution. Identified peaks are marked in different colors for the various methods.
Supplementary Figure 3 Concordance of cis and trans interactions called by the various tools (Jaccard Index).
a) Box plots of the Jaccard Index for concordance of cis (upper panels) and trans (lower panels) interaction calls between sample replicates in any dataset (intra-dataset concordance). Jaccard Index was not calculated for GOTHiC in Dixon 2015 (see Supplementary Note 6) and for HIPPIE in cis interactions of Lieberman-Aiden (see Supplementary Figure 2). b) Stacked bar plot for the number of pairwise comparisons of cis interactions between replicates stratified by significance. The y-axis scale depends on the number of pairwise comparison per dataset. Bars are colored according to the tool if the comparisons have a Jaccard Index p-value ≤0.001 and with shades of grey for comparisons with Jaccard Index p-value >0.001. Empirical p-values were estimated with random permutations of interactions. Briefly, for each dataset, cell type, and data analysis method, we defined, for each sample, a random set of cis interactions by keeping constant the sample-specific number of interactions and the sample-specific distribution of distances between anchoring points. The first of the two anchoring points for each interaction was randomly selected from the pool of detectable anchoring points, defined as any genomic bin that was called as anchoring point in any sample from the same dataset and cell type. The second anchoring point was randomly defined by sampling from the observed distribution of anchoring point distances. The resulting sets of random interactions were then used to compute random Jaccard Index values in pairwise comparisons. The random sampling of interactions was repeated 1000 times to obtain a null distribution of randomly expected Jaccard indexes for each pairwise comparison. The empirical p-value is estimated as the probability of observing a random Jaccard Index value larger than or equal to the observed one. Almost all of the observed Jaccard indexes in the pairwise comparisons are significantly larger than expected by chance. Stacked bars lower than the maximum value are used for samples including one or more replicates with no detected interactions.
Supplementary Figure 4 Concordance of cis interactions called in Rao dataset and overlap coefficients of cis and trans interactions in all datasets.
a) Box plots of Jaccard Index of all (left) and top 1000 (right) cis interaction calls between replicates A1, A2, A5, B1, and B2 of IMR90 samples in Rao dataset. The top 1,000 interactions were defined based on the False Discovery Rate (FDR) for HiCCUPS, GOTHiC, and Fit-Hi-C, on the p-value for HOMER and HIPPIE, and using the enrichment score in diffHic. b) Scatter plot and linear interpolation of average Jaccard Index (y-axis) versus average number of read pairs (x-axis in log scale) in Rao GM12878 replicates stratified by number of reads (see Online Methods). The plot shows that for HiCCUPS and GOTHiC the Jaccard Index has a stronger increase in pairwise comparisons between samples in groups with larger number of reads. c) Box plots of the overlap coefficient for concordance of cis (upper panels) and trans (lower panels) interaction calls between sample replicates in any dataset (intra-dataset concordance). The overlap coefficient is measured as the size of the common set of interactions in a pairwise comparison, divided by the size of the smallest between the two compared sets. The overlap coefficient was not calculated for GOTHiC in Dixon 2015 (see Supplementary Note 6) and for HIPPIE in cis interactions of Lieberman-Aiden (see Supplementary Figure 2).
Supplementary Figure 5 Concordance of cis interactions called in Rao GM12878 and in Lieberman-Aiden GM06990 processed with different restriction enzymes.
a) Heatmaps of the Jaccard Index of cis interaction calls between replicates of Rao GM12878 cell line processed with DpnII (green sidebar) or MboI (purple sidebar) restriction enzymes. The dashed box contains the Jaccard Index of each pair of DpnII ‑ MboI processed replicates. For each enzyme, replicates are ordered according to decreasing sequencing depth. The color scale is the same for all heatmaps. b) Box plots of the Jaccard Index of cis interaction calls between all pairs of DpnII ‑ MboI Rao GM12878 processed replicates. c) Box plots of the Jaccard Index of cis interaction calls between all pairs of NcoI ‑ HindIII Lieberman-Aiden GM06990 processed replicates. Jaccard Index was not calculated for HIPPIE (see Supplementary Figure 2).
Supplementary Figure 6 Concordance of cis interactions called in replicates of IMR90 from Rao and Jin datasets.
a) Heatmaps of the Jaccard Index of cis interaction calls between IMR90 replicates from Rao (green sidebar) and Jin (purple sidebar) datasets. Replicates from the two datasets differ in terms of restriction enzyme and Hi-C protocol (4bp cutter MboI and in-situ for Rao; 6bp cutter HindIII and dilution for Jin). The dashed box contains the Jaccard Index of each pair of Rao IMR90 ‑ Jin IMR90 replicates (inter-dataset concordance). For each dataset, replicates are ordered according to decreasing sequencing depth. The color scale is the same for all heatmaps. b) Box plots of the Jaccard Index of cis interaction calls between all pairs of Rao IMR90 ‑ Jin IMR90 replicates.
Supplementary Figure 7 Absolute number (datasets at 5kb and 40kb) and proportion (datasets at 40kb resolution) of cis interactions classified on the base of the chromatin states and percentage of true-positive and true-negative interactions recalled by each tool.
a) Absolute number of cis interactions classified on the base of the chromatin states at their anchoring points as promoter-enhancer (upper), heterochromatin/quiescent to heterochromatin/quiescent (middle), and less expected (lower) in all datasets at 5kb (data not shown for interactions classified as other combinations of chromatin states). With the exception of Jin H1-hESC (that contains a single replicate), only cis interactions conserved in at least 2 replicates within each dataset were classified using the chromatin states (Supplementary Table 4). b) Proportion (left) and absolute number (right) of cis interactions classified as in a) in all datasets at 40kb (data not shown for interactions classified as other combinations of chromatin states). With the exception of Sexton dataset (that contains a single replicate), only cis interactions conserved in at least 2 replicates within each dataset were classified using the chromatin states. c) Percentage of true-positive interactions (%TP) from 5C data of Sanyal et al. (see Supplementary Table 7) recalled, in each replicate of Rao GM12878 dataset (5kb resolution), by each method as a function of the total number of called cis interactions (x-axis in log scale). We used data from Rao GM12878 since Rao dataset contained the largest number of replicates for GM12878 cell line and GM12878 was characterized by a large number of known true positives. d) Performances in the identification of true negative validated evidences of cis interactions. Each column represents the comparison between a list of true negatives and the interactions called by each method in each dataset. The dot size is proportional to the percentage of recalled true negatives and the dot color accounts for the number of total called interactions. The validation technique and the name of true negative lists are displayed on top. The datasets used to call interactions are at the bottom. Datasets at 40 kb resolution are shaded in grey. True-negative interactions were searched among cis interactions conserved in at least 2 replicates within each dataset, with the exception of Jin H1-hESC (which contains a single replicate). GOTHiC was not applied to Dixon 2015 (see Supplementary Note 6).
Supplementary Figure 8 Simulation results for interaction callers.
a) Average number of cis interactions called by each method as a function of the base interaction strength without the additional fixed constant (Kinteractions, see Supplementary note 3). The number of true interactions (1,000) is shown as a dashed line. Data are shown as mean±standard error of the mean (SEM). Similar results were obtained using the additional fixed constant (data not shown). b) Boxplot of average distances between anchoring points in cis interactions (log scale) in 5 replicates generated at a base interaction strength equal to 4 times the baseline of simulated TADs. c) Heatmap of the contact matrix generated with base interaction strength equal to 2 times the baseline of simulated TADs (simulated chr:0-8,000,000). True simulated interaction peaks are in green, identified peaks are marked in different colors for the various methods. d) True positive rate (sensitivity) as a function of the base interaction strength with (dashed line) and without (solid line) the Kinteractions constant. Data are shown as mean±standard error of the mean. e) False Discovery Rate (1-precision) as a function of the base interaction strength with (dashed line) and without (solid line) the Kinteractions constant. Data are shown as mean±standard error of the mean.
Supplementary Figure 9 Intra-dataset concordance of TAD boundaries.
a) Box plots of the Jaccard Index for concordance of TAD boundaries between pairs of sample replicates in each dataset (intra-dataset). b) Scatter plot and linear interpolation of average Jaccard Index (y-axis) versus average number of read pairs (x-axis in log scale) in Rao GM12878 replicates stratified by number of reads (see Online Methods). The plot shows that for all tools the Jaccard Index has a tendency to increase in pairwise comparisons between samples in groups with larger number of reads. c) Box plots of the overlap coefficient for concordance of TAD boundaries between sample replicates in any dataset (intra-dataset concordance). The overlap coefficient is measured as the size of the common set of TAD boundaries in a pairwise comparison, divided by the size of the smallest between the two compared sets.
Supplementary Figure 10 Concordance of TAD boundaries in replicates of Rao GM12878 and in Lieberman-Aiden GM06990 processed with different restriction enzymes.
a) Heatmaps of the Jaccard Index of TAD boundaries concordance between replicates of Rao GM12878 cell line processed with DpnII (green sidebar) and MboI (purple sidebar) restriction enzymes. The dashed box contains the Jaccard Index of each pair of DpnII ‑ MboI processed replicates. For each enzyme, replicates are ordered according to decreasing number of reads retained after filtering. The color scale is the same for all heatmaps. b) Box plots of the Jaccard Index of TAD boundaries between all pairs of DpnII ‑ MboI Rao GM12878 processed replicates. c) Box plots of the Jaccard Index of TAD boundaries between all pairs of NcoI ‑ HindIII Lieberman-Aiden GM06990 processed replicates.
Supplementary Figure 11 Concordance of TAD boundaries in replicates of IMR90 from Rao and Jin datasets.
a) Heatmaps of the Jaccard Index of TAD boundaries concordance between IMR90 replicates from Rao (green sidebar) and Jin (purple sidebar) datasets. Replicates from the two datasets differ in terms of restriction enzyme and Hi-C protocol (4bp cutter MboI and in-situ for Rao; 6bp cutter HindIII and dilution for Jin). The dashed box contains the Jaccard Index of each pair of Rao IMR90 ‑ Jin IMR90 replicates (inter-dataset concordance). For each dataset, replicates are ordered according to decreasing number of reads retained after filtering. The color scale is the same for all heatmaps. b) Box plots of the Jaccard Index of TAD boundaries between all pairs of Rao IMR90 ‑ Jin IMR90 replicates.
Supplementary Figure 12 Enrichment of insulator binding around the TAD boundaries.
a) Enrichment of CTCF binding (ChIP-seq peaks) in a window of 1Mb around the TAD boundaries (all datasets). With the exception of Sexton dataset (that contains a single replicate), only TAD boundaries conserved in at least 2 replicates within each dataset were used to calculate the CTCF binding enrichment. The enrichment for Arrowhead in Dixon 2012 H1-hESC was not calculated since Arrowhead found only one conserved TAD boundary in this dataset (see Supplementary Table 4). The less sharp enrichment of CTCF peaks at TAD boundaries identified by InsulationScore may be partly explained by the observation reported in Crane et al. (Nature 2015) that the boundary position determined by InsulationScore should be defined as a zone around the insulation minimum rather than as a single bin position. b) Enrichment of BEAF32 binding (ChIP-seq peaks) in a window of 1Mb around the TAD boundaries (Sexton dataset).
Supplementary Figure 13 Simulation results for TAD callers.
a) Average number of TADs called by each method as a function of the simulated noise level. The number of true TADs (171) is shown as a dashed line. Data are shown as mean±standard error of the mean (SEM). Arrowhead identified only 1 TAD in 1 simulated matrix and thus results for this tool are not reported here. b) Boxplot of median sizes for TADs called by the various methods in 5 replicates generated at a noise level equal to the 12% of the total number of data points of the simulated matrices. The 1st and 3rd quartile of the distribution of median true TAD sizes are shown as dashed lines. c) True positive rate in the identification of TAD boundaries as a function of the noise level (sensitivity). Data are shown as mean±standard error of the mean. d) False Discovery Rate (1-precision) in the identification of TAD boundaries as a function of the noise level. Data are shown as mean±standard error of the mean. e) Heatmaps of the contact matrix generated with nested TADs at a noise level equal to the 4% of the total number of data points of the simulated matrices (simulated chr:127,000,000-137,000,000). True simulated nested TADs are in green, called TADs by are marked in different colors for each method. f) Same as in c) for nested TADs. g) Same as in d) for nested TADs.
Supplementary Figure 14 Identification of chromatin interactions using a common preprocessing.
We applied hiclib as a common preprocessing procedure to align and filter reads from Dixon2012 IMR90 and Jin IMR90. These data were then used as input to all tools, with the exception of HIPPIE, for which it is not possible to disentangle preprocessing and downstream analysis. Normalization and downstream analysis were performed using each tool proprietary procedures. We used Juicer Tools Pre to convert hiclib output into the.hic input file for HiCCUPS. a) Percentage of aligned read pairs (alignment rate) for all approaches, including hiclib iterative mapping. Data are shown as mean±standard error of the mean. b) Percentage of mapped reads retained after filtering (fraction of usable reads) for all tools, including hiclib. Data are shown as mean±standard error of the mean. c) Scatter plot of total number of cis interactions called by each method versus the number of reads retained by the filtering steps in Jin IMR90 dataset. Different points represent sample replicates analyzed using hiclib common preprocessing (filled dots) or the preprocessing of each tool (open circles). Linear interpolation (of log transformed data) is shown as solid line for hiclib common preprocessing and as dashed line for each tool preprocessing. d) Box plots of the Jaccard Index of cis interaction calls between sample replicates in Dixon2012 IMR90 and Jin IMR90 commonly preprocessed using hiclib (left panel) or using each single tool (right panel). e) Proportion of cis interactions classified as promoter-enhancer in Dixon2012 IMR90 and Jin IMR90 commonly preprocessed using hiclib (left panel) or using each single tool (right panel). f) Overlap coefficient of cis interactions called preprocessing the data using hiclib or the alignment and filtering steps of each single tool in every sample of Dixon2012 IMR90 and Jin IMR90 (n=8).
Supplementary Figure 15 Running time of interaction and TAD callers.
a) Time required by the various methods to perform alignment, reads pairing and PCR duplicates removal, other filtering, and normalization-downstream analysis for calling interactions in single replicates at different resolutions (replicate B of Dixon IMR90 at 40kb and replicate A5 of Rao IMR90 at 5kb; n=2 samples). The analyses were run on a single CPU and on a GPU for HiCCUPS (Online Methods). For GOTHiC, HOMER, and Fit-Hi-C the alignment time is relative to Bowtie. The time for reads pairing and PCR duplicates removal and other filtering of Fit-Hi-C corresponds to that of GOTHiC. b) Time required by the various methods to perform alignment, preprocessing (pairing, filtering, and normalization) and downstream analysis for TAD calling in replicates B of Dixon IMR90 and A5 of Rao IMR90 (n=2 samples). Alignment and preprocessing time are the same for all tools since all methods have been applied to a matrix generated by hicpipe. For TADbit, the time of downstream analysis also accounts for the normalization step. Both samples were analyzed at 40kb resolution. However, Rao IMR90 replicate A5 required a higher preprocessing running time due to the large number of restriction fragments generated by the 4bp cutter restriction enzyme.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–10, Supplementary Notes 1–7 and Supplementary Figures 1–15
Source data
Rights and permissions
About this article
Cite this article
Forcato, M., Nicoletti, C., Pal, K. et al. Comparison of computational methods for Hi-C data analysis. Nat Methods 14, 679–685 (2017). https://doi.org/10.1038/nmeth.4325
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4325
This article is cited by
-
Computational methods for analysing multiscale 3D genome organization
Nature Reviews Genetics (2024)
-
BRWD1 orchestrates small pre-B cell chromatin topology by converting static to dynamic cohesin
Nature Immunology (2024)
-
Regulation and function of transposable elements in cancer genomes
Cellular and Molecular Life Sciences (2024)
-
SnapFISH: a computational pipeline to identify chromatin loops from multiplexed DNA FISH data
Nature Communications (2023)
-
Context-dependent perturbations in chromatin folding and the transcriptome by cohesin and related factors
Nature Communications (2023)