Abstract
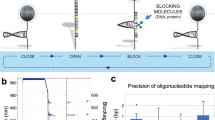
High-throughput, low-cost DNA sequencing has emerged as one of the challenges of the postgenomic era. Here we present the proof of concept for a single-molecule platform that allows DNA identification and sequencing. In contrast to most present methods, our scheme is not based on the detection of the fluorescent nucleotides but on DNA hairpin length. By pulling on magnetic beads tethered by a DNA hairpin to the surface, the molecule can be unzipped. In this open state it can hybridize with complementary oligonucleotides, which transiently block the hairpin rezipping when the pulling force is reduced. By measuring from the surface to the bead of a blocked hairpin, one can determine the position of the hybrid along the molecule with nearly single-base precision. Our approach can be used to identify a DNA fragment of known sequence in a mix of various fragments and to sequence an unknown DNA fragment by hybridization or ligation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Braslavsky, I., Hebert, B., Kartalov, E. & Quake, S.R. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. USA 100, 3960–3964 (2003).
Pushkarev, D., Neff, N.F. & Quake, S.R. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 27, 847–850 (2009).
Pihlak, A. et al. Rapid genome sequencing with short universal tiling probes. Nat. Biotechnol. 26, 676–684 (2008).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Shendure, J., Mitra, R.D., Varma, C. & Church, G.M. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 5, 335–344 (2004).
Metzker, M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46 (2010).
Fuller, C.W. et al. The challenges of sequencing by synthesis. Nat. Biotechnol. 27, 1013–1023 (2009).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Greenleaf, W.J. & Block, S.M. Single-molecule, motion-based DNA sequencing using RNA polymerase. Science 313, 801 (2006).
Munroe, D.J. & Harris, T.J.R. Third-generation sequencing fireworks at marco island. Nat. Biotechnol. 28, 426–428 (2010).
Clarke, J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009).
Treffer, R. & Deckert, V. Recent advances in single-molecule sequencing. Curr. Opin. Biotechnol. 21, 4–11 (2010).
Husale, S., Persson, H.H.J. & Sahin, O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature 462, 1075–1078 (2009).
Clark, M.D. et al. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome Res. 11, 1594–1602 (2001).
Herwig, R. et al. Information theoretical probe selection for hybridisation experiments. Bioinformatics 16, 890–898 (2000).
Guerasimova, A. et al. New tools for oligonucleotide fingerprinting. Biotechniques 31, 490–495 (2001).
Gosse, C. & Croquette, V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 82, 3314–3329 (2002).
Brower-Toland, B.D. et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA 99, 1960–1965 (2002).
Strick, T.R., Allemand, J., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996).
Essevaz-Roulet, B., Bockelmann, U. & Heslot, F. Mechanical separation of the complementary strands of DNA. Proc. Natl. Acad. Sci. USA 94, 11935–11940 (1997).
McNally, B. et al. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 10, 2237–2244 (2010).
Mir, K.U., Qi, H., Salata, O. & Scozzafava, G. Sequencing by cyclic ligation and cleavage (CycLiC) directly on a microarray captured template. Nucleic Acids Res. 37, e5 (2009).
Manosas, M., Spiering, M.M., Zhuang, Z., Benkovic, S.J. & Croquette, V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat. Chem. Biol. 5, 904–912 (2009).
Kim, K. & Saleh, O.A. A high-resolution magnetic tweezer for single-molecule measurements. Nucleic Acids Res. 37, e136 (2009).
De Vlaminck, I. et al. Highly parallel magnetic tweezers by targeted DNA tethering. Nano Lett. 11, 5489–5493 (2011).
Singh-Zocchi, M., Dixit, S., Ivanov, V. & Zocchi, G. Single-molecule detection of DNA hybridization. Proc. Natl. Acad. Sci. USA 100, 7605–7610 (2003).
Melchior, W.B. & Von Hippel, P.H. Jr. Alteration of the relative stability of dA - dT and dG* dC base pairs in DNA. Proc. Natl. Acad. Sci. USA 70, 298–302 (1973).
Acknowledgements
We acknowledge useful suggestions by M. Volovitch, T. Lionnet and K. Neuman. This work was supported by an ERA-MolMachines grant (to D.B.), a Human Frontier Science Program grant (RGP003/2007 to V.C. and S.J.B.) and the European Research Council grant 'MagRepS' 267862 (to V.C., F.D., M.M. and S.J.B.). We thank J. Quintas for providing mechanical expertise on the instrument.
Author information
Authors and Affiliations
Contributions
J.-F.A. and V.C. designed the apparatus; F.D., S.J.B., M.M., M.M.S. and V.C. discussed the T4 system that led to the concept of sequencing; F.D. and M.M. performed experiments on magnetic tweezers; F.D. and M.M.S. prepared DNA hairpins; F.D., D.B. and V.C. performed hybridization and ligation assays; F.D. and V.C. analyzed data; F.D., M.M., M.M.S., S.J.B., J.-F.A., D.B. and V.C. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.B., J-F.A. and V.C. are collaborating with the company PicoTwist that manufactures magnetic tweezers. F.D., M.M., D.B., J-F.A. and V.C. have three patent applications on the work described here (EP 10305563.8, EP 10305564.6 and EP 11306743).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Discussion, Supplementary Note 1 (PDF 1441 kb)
Rights and permissions
About this article
Cite this article
Ding, F., Manosas, M., Spiering, M. et al. Single-molecule mechanical identification and sequencing. Nat Methods 9, 367–372 (2012). https://doi.org/10.1038/nmeth.1925
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1925
This article is cited by
-
Detection of genetic variation and base modifications at base-pair resolution on both DNA and RNA
Communications Biology (2021)
-
Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis
Nature Communications (2018)
-
Unfolding of a ClC chloride transporter retains memory of its evolutionary history
Nature Chemical Biology (2018)
-
A rapid and practical technique for real-time monitoring of biomolecular interactions using mechanical responses of macromolecules
Scientific Reports (2016)
-
Camera-based three-dimensional real-time particle tracking at kHz rates and Ångström accuracy
Nature Communications (2015)