Abstract
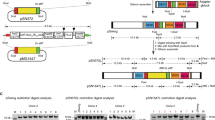
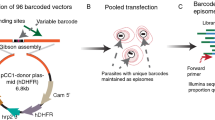
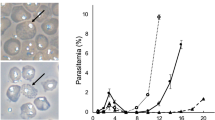
In malaria parasites, the systematic experimental validation of drug and vaccine targets by reverse genetics is constrained by the inefficiency of homologous recombination and by the difficulty of manipulating adenine and thymine (A+T)-rich DNA of most Plasmodium species in Escherichia coli. We overcame these roadblocks by creating a high-integrity library of Plasmodium berghei genomic DNA (>77% A+T content) in a bacteriophage N15–based vector that can be modified efficiently using the lambda Red method of recombineering. We built a pipeline for generating P. berghei genetic modification vectors at genome scale in serial liquid cultures on 96-well plates. Vectors have long homology arms, which increase recombination frequency up to tenfold over conventional designs. The feasibility of efficient genetic modification at scale will stimulate collaborative, genome-wide knockout and tagging programs for P. berghei.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Crabb, B.S. & Cowman, A.F. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 93, 7289–7294 (1996).
van Dijk, M.R., Janse, C.J. & Waters, A.P. Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science 271, 662–665 (1996).
Wu, Y., Kirkman, L.A. & Wellems, T.E. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc. Natl. Acad. Sci. USA 93, 1130–1134 (1996).
Tewari, R. et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe 8, 377–387 (2010).
Maier, A.G. et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 (2008).
Janse, C.J., Ramesar, J. & Waters, A.P. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356 (2006).
Hasty, P., Rivera-Perez, J. & Bradley, A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11, 5586–5591 (1991).
Shulman, M.J., Nissen, L. & Collins, C. Homologous recombination in hybridoma cells: dependence on time and fragment length. Mol. Cell. Biol. 10, 4466–4472 (1990).
Gardner, M.J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Hall, N. et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 (2005).
Ejsmont, R.K., Sarov, M., Winkler, S., Lipinski, K.A. & Tomancak, P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6, 435–437 (2009).
Poser, I. et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 (2008).
Sarov, M. et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3, 839–844 (2006).
Ravin, N.V. & Ravin, V.K. Use of a linear multicopy vector based on the mini-replicon of temperate coliphage N15 for cloning DNA with abnormal secondary structures. Nucleic Acids Res. 27, e13 (1999).
Ravin, N.V., Kuprianov, V.V., Gilcrease, E.B. & Casjens, S.R. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 31, 6552–6560 (2003).
Godiska, R. et al. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res. 38, e88 (2010).
Zhang, Y., Buchholz, F., Muyrers, J.P. & Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998).
Wang, J. et al. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 32, 43–53 (2006).
Gubbels, M.J. et al. Forward genetic analysis of the apicomplexan cell division cycle in Toxoplasma gondii. PLoS Pathog. 4, e36 (2008).
Brooks, C.F. et al. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe 7, 62–73 (2010).
Skarnes, W.C. et al. A conditional knockout resource for genome-wide analysis of mouse gene function. Nature 474, 337–342 (2011).
Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009).
Braks, J.A., Franke-Fayard, B., Kroeze, H., Janse, C.J. & Waters, A.P. Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 34, e39 (2006).
Menard, R. & Janse, C.J. Gene targeting in malaria parasites. Methods 13, 148–157 (1997).
Le Roch, K.G. et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 (2003).
Tremp, A.Z. & Dessens, J.T. Malaria IMC1 membrane skeleton proteins operate autonomously and participate in motility independently of cell shape. J. Biol. Chem. 286, 5383–5391 (2011).
Moon, R.W. et al. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog. 5, e1000599 (2009).
Janse, C.J., Franke-Fayard, B. & Waters, A.P. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat. Protoc. 1, 614–623 (2006).
Rutherford, K. et al. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 (2000).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Assefa, S., Keane, T.M., Otto, T.D., Newbold, C. & Berriman, M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25, 1968–1969 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Janse, C.J. et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 145, 60–70 (2006).
Acknowledgements
We thank F. Stewart (Technische Universität Dresden) for sharing plasmids pSC101gbdA and pR6K attR1-zeo-PheS-attR2, C. Janse (Leiden University Medical Center) and A. Waters (University of Glasgow) for sharing P. berghei clones RmGM-7 and RmGM-29 and plasmid pL0035, and J. Warren and H. Wang for helping to create a Distributed Annotation System source of clone data for PlasmoDB. This work was supported by the Wellcome Trust (WT089085/Z/09/Z), the Medical Research Council (G0501670) and the European Virtual Institute for Malaria Research Network of Excellence (242095).
Author information
Authors and Affiliations
Contributions
J.C.R and O.B. initiated and directed the research. C.P., A.P., B.R., W.S., J.C.R. and O.B. designed experiments. F.S., T.D.O., M.A.Q. and A.P. generated, sequenced, mapped and quality controlled the PbG01 library. C.P. and B.A. carried out experiments to develop the recombineering pipeline. C.P., M.B. and K.V. carried out experiments to validate the vectors. C.P. and O.B. wrote the manuscript. All authors analyzed data and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Tables 1–2, Supplementary Protocols 1–3 (PDF 554 kb)
Rights and permissions
About this article
Cite this article
Pfander, C., Anar, B., Schwach, F. et al. A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat Methods 8, 1078–1082 (2011). https://doi.org/10.1038/nmeth.1742
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1742
This article is cited by
-
The Skp1-Cullin1-FBXO1 complex is a pleiotropic regulator required for the formation of gametes and motile forms in Plasmodium berghei
Nature Communications (2023)
-
Plasmodium ARK2 and EB1 drive unconventional spindle dynamics, during chromosome segregation in sexual transmission stages
Nature Communications (2023)
-
Fast and efficient generation of a full-length balancer chromosome by a single Cre/loxP recombination event
Mammalian Genome (2022)
-
Plasmodium vinckei genomes provide insights into the pan-genome and evolution of rodent malaria parasites
BMC Biology (2021)
-
Limited Plasmodium sporozoite gliding motility in the absence of TRAP family adhesins
Malaria Journal (2021)