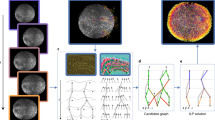
Abstract
Understanding how stem and progenitor cells choose between alternative cell fates is a major challenge in developmental biology. Efforts to tackle this problem have been hampered by the scarcity of markers that can be used to predict cell division outcomes. Here we present a computational method, based on algorithmic information theory, to analyze dynamic features of living cells over time. Using this method, we asked whether rat retinal progenitor cells (RPCs) display characteristic phenotypes before undergoing mitosis that could foretell their fate. We predicted whether RPCs will undergo a self-renewing or terminal division with 99% accuracy, or whether they will produce two photoreceptors or another combination of offspring with 87% accuracy. Our implementation can segment, track and generate predictions for 40 cells simultaneously on a standard computer at 5 min per frame. This method could be used to isolate cell populations with specific developmental potential, enabling previously impossible investigations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cayouette, M., Poggi, L. & Harris, W.A. Lineage in the vertebrate retina. Trends Neurosci. 29, 563–570 (2006).
Cayouette, M., Barres, B.A. & Raff, M. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron 40, 897–904 (2003).
Godinho, L. et al. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron 56, 597–603 (2007).
Mu, X. et al. Ganglion cells are required for normal progenitor-cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Curr. Biol. 15, 525–530 (2005).
Poggi, L., Vitorino, M., Masai, I. & Harris, W.A. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J. Cell Biol. 171, 991–999 (2005).
Diaz, E. et al. Analysis of gene expression in the developing mouse retina. Proc. Natl. Acad. Sci. USA 100, 5491–5496 (2003).
Dorrell, M.I., Aguilar, E., Weber, C. & Friedlander, M. Global gene expression analysis of the developing postnatal mouse retina. Invest. Ophthalmol. Vis. Sci. 45, 1009–1019 (2004).
Livesey, F.J., Young, T.L. & Cepko, C.L. An analysis of the gene expression program of mammalian neural progenitor cells. Proc. Natl. Acad. Sci. USA 101, 1374–1379 (2004).
Mu, X. et al. Gene expression in the developing mouse retina by EST sequencing and microarray analysis. Nucleic Acids Res. 29, 4983–4993 (2001).
Trimarchi, J.M., Stadler, M.B. & Cepko, C.L. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One 3, e1588 (2008).
Tietjen, I. et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron 38, 161–175 (2003).
Jessell, T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29 (2000).
Cohen, A.R., Bjornsson, C.S., Temple, S., Banker, G. & Roysam, B. Automatic summarization of changes in biological image sequences using algorithmic information theory. IEEE Trans. Pattern Anal. Mach. Intell. 31, 1386–1403 (2009).
Kamvar, S.D., Klein, D. & Manning, C.D. Spectral learning. International Joint Conference of Artificial Intelligence (2003).
Baye, L.M. & Link, B. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci. 27, 10143–10152 (2007).
Cilibrasi, R. & Vitanyi, P.M.B. Clustering by compression. IEEE Trans. Inf. Theory 51, 1523–1545 (2005).
Witten, I.H. & Frank, E. Data Mining: Practical Machine Learning Tools and Techniques (Morgan Kaufmann, 2005).
Chen, Y., Ladi, E., Herzmark, P., Robey, E. & Roysam, B. Automated 5-D analysis of cell migration and interaction in the thymic cortex from time-lapse sequences of 3-D multi-channel multi-photon images. J. Immunol. Methods 340, 65–80 (2009).
Barres, B.A. et al. Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46 (1992).
Barres, B.A., Lazar, M.A. & Raff, M.C. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development 120, 1097–1108 (1994).
Soille, P. Morphological Image Analysis: Principles and Applications (Springer-Verlag, 1999).
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991).
Lin, J., Keogh, E., Lonardi, S. & Chiu, B. A symbolic representation of time series, with implications for streaming algorithms. Data Min. Knowl. Discov. 15, 107–144 (2007).
Ng, A.Y., Jordan, M. & Weiss, Y. On Spectral Clustering: Analysis and an algorithm. Adv. Neural Inf. Process. Syst 14, 849–856 (2001).
Al-Kofahi, O. et al. Automated cell lineage tracing: a high-throughput method to analyze cell proliferative behavior developed using mouse neural stem cells. Cell Cycle 5, 327–335 (2006).
Debeir, O., Van Ham, P., Kiss, R. & Decaestecker, C. Tracking of migrating cells under phase-contrast video microscopy with combined mean-shift processes. IEEE Trans. Med. Imaging 24, 697–711 (2005).
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008).
Li, K. et al. Cell population tracking and lineage construction with spatiotemporal context. Med. Image Anal. 12, 546–566 (2008).
Meijering, E., Smal, I. & Danuser, G. Tracking in molecular bioimaging. IEEE Signal Process. Mag. 23, 46–53 (2006).
Bennett, C.H., Gacs, P., Ming, L., Vitanyi, M.B. & Zurek, W.H. Information distance. IEEE Trans. Inf. Theory 44, 1407–1423 (1998).
Li, M. & Vitanyi, P.M.B. An Introduction to Kolmogorov Complexity and Its Applications (Springer Verlag, New York, 1997).
Li, M., Chen, X., Li, X., Ma, B. & Vitanyi, P.M.B. The similarity metric. IEEE Trans. Inf. Theory 50, 3250–3264 (2004).
Cebrian, M., Alfonseca, M. & Ortega, A. The normalized compression distance is resistant to noise. IEEE Trans. Inf. Theory 53, 1895–1900 (2007).
Keogh, E., Lonardi, S. & Ratanamahatana, C.A. Towards parameter-free data mining. in Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM Press, Seattle, 2004).
Rissanen, J. Stochastic Complexity in Statistical Inquiry (World Scientific, Singapore, 1989).
Grünwald, P., Myung, I.J. & Pitt, M. Advances in Minimum Description Length: Theory and Applications (MIT Press, 2005).
Acknowledgements
The computational aspects of this work were supported by the Center for Subsurface Sensing and Imaging Systems (US National Science Foundation; EEC-9986821), by the Rensselaer Polytechnic Institute and by the University of Wisconsin–Milwaukee. We thank A. Todorski and the staff at Rensselaer's Computational Center for Nanotechnology Innovations supercomputing center; S. Temple for helpful comments and feedback, and on collaborations leading up to the present work; M. Kmita and J. Chan for insightful comments on the manuscript; A. Daigneault for expert technical assistance with the animal colony; and C. Jolicoeur for help with the oligodendrocyte precursor cell culture. This work was supported by grants from the Canadian Institutes of Health Research and the Foundation Fighting Blindness-Canada (to M.C.). M.C. is supported by the Canadian Institutes of Health Research New Investigator program and the W.K. Stell scholarship from the Foundation Fighting Blindness, Canada.
Author information
Authors and Affiliations
Contributions
A.R.C. and B.R. developed the computational methods; F.L.A.F.G. and M.C. proposed the initial hypothesis and developed the cell culture, immunostaining and imaging methods. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–3 (PDF 917 kb)
Supplementary Movie 1
Retinal progenitor cell undergoing a self-renewing division (Fig. 2). A RPC divides to give rise to a photoreceptor and another RPC at the first and second division (self-renewing) and finally undergoes a terminal division to give rise to two photoreceptors. (MOV 899 kb)
Supplementary Movie 2
Retinal progenitor cell undergoing a terminal division (Fig. 3). A RPC divides to give rise to a photoreceptor and an amacrine cell. (MOV 2637 kb)
Supplementary Movie 3
Example of a retinal progenitor cell segmentation and tracking results. (MOV 986 kb)
Supplementary Movie 4
Oligodendrocyte precursor cell undergoing a terminal division (Supplementary Figure 1). An OPC divides to give rise to two oligodendrocytes. (MOV 2834 kb)
Supplementary Movie 5
Oligodendrocyte precursor cell undergoing a self-renewing division (Supplementary Figure 2). An OPC divides to give rise to another OPC and an oligodendrocyte (self-renewing). (MOV 4085 kb)
Supplementary Software
Source code for the AITP software. (ZIP 28619 kb)
Rights and permissions
About this article
Cite this article
Cohen, A., Gomes, F., Roysam, B. et al. Computational prediction of neural progenitor cell fates. Nat Methods 7, 213–218 (2010). https://doi.org/10.1038/nmeth.1424
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1424
This article is cited by
-
Web Similarity in Sets of Search Terms Using Database Queries
SN Computer Science (2020)
-
ATLANTIS - Attractor Landscape Analysis Toolbox for Cell Fate Discovery and Reprogramming
Scientific Reports (2018)
-
Machine learning applications in cell image analysis
Immunology & Cell Biology (2017)
-
Prospective identification of hematopoietic lineage choice by deep learning
Nature Methods (2017)
-
Resolution-enhanced Fourier ptychographic microscopy based on high-numerical-aperture illuminations
Scientific Reports (2017)