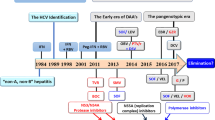
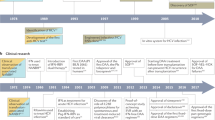
Abstract
More than two decades of intense research has provided a detailed understanding of hepatitis C virus (HCV), which chronically infects 2% of the world's population. This effort has paved the way for the development of antiviral compounds to spare patients from life-threatening liver disease. An exciting new era in HCV therapy dawned with the recent approval of two viral protease inhibitors, used in combination with pegylated interferon-α and ribavirin; however, this is just the beginning. Multiple classes of antivirals with distinct targets promise highly efficient combinations, and interferon-free regimens with short treatment duration and fewer side effects are the future of HCV therapy. Ongoing and future trials will determine the best antiviral combinations and whether the current seemingly rich pipeline is sufficient for successful treatment of all patients in the face of major challenges, such as HCV diversity, viral resistance, the influence of host genetics, advanced liver disease and other co-morbidities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Thomas, D.L. Nat. Med. 19, 850–858 (2013).
Feld, J.J. & Hoofnagle, J.H. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436, 967–972 (2005).
Manns, M.P., Wedemeyer, H. & Cornberg, M. Treating viral hepatitis C: Efficacy, side effects, and complications. Gut 55, 1350–1359 (2006).
Jacobson, I.M. et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364, 2405–2416 (2011).
Poordad, F. et al. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364, 1195–1206 (2011).
Sarrazin, C., Hezode, C., Zeuzem, S. & Pawlotsky, J.M. Antiviral strategies in hepatitis C virus infection. J. Hepatol. 56 (suppl. 1), S88–S100 (2012).
Liang, T.J. Nat. Med. 19, 869–878 (2013).
Choo, Q.L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989).
Alter, H.J. et al. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N. Engl. J. Med. 321, 1494–1500 (1989).
Kuo, G. et al. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244, 362–364 (1989).
Bukh, J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 142, 1279–1287.e3 (2012).
Mercer, D.F. et al. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7, 927–933 (2001).
Dorner, M. et al. A genetically humanized mouse model for hepatitis C virus infection. Nature 474, 208–211 (2011).
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999).
Bartosch, B., Dubuisson, J. & Cosset, F.L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197, 633–642 (2003).
Hsu, M. et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100, 7271–7276 (2003).
Wakita, T. et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11, 791–796 (2005).
Lindenbach, B.D. et al. Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 (2005).
Zhong, J. et al. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102, 9294–9299 (2005).
Kapoor, A. et al. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. USA 108, 11608–11613 (2011).
Kapoor, A. et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio 4, e00216–e00213 (2013).
Quan, P.L. et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 110, 8194–8199 (2013).
Hoffman, B. & Liu, Q. Hepatitis C viral protein translation: Mechanisms and implications in developing antivirals. Liver Int. 31, 1449–1467 (2011).
Gottwein, J.M. & Bukh, J. Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems. Adv. Virus Res. 71, 51–133 (2008).
Bartenschlager, R., Penin, F., Lohmann, V. & Andre, P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19, 95–103 (2011).
Negro, F. Steatosis and insulin resistance in response to treatment of chronic hepatitis C. J. Viral Hepat. 19 (suppl. 1), 42–47 (2012).
van der Meer, A.J. et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. J. Am. Med. Assoc. 308, 2584–2593 (2012).
Kau, A., Vermehren, J. & Sarrazin, C. Treatment predictors of a sustained virologic response in hepatitis B and C. J. Hepatol. 49, 634–651 (2008).
Horner, S.M. & Gale, M. Jr. Nat. Med. 19, 879–888 (2013).
Gastaminza, P. et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84, 10999–11009 (2010).
Bassendine, M.F. et al. Hcv and the hepatic lipid pathway as a potential treatment target. J. Hepatol. 55, 1428–1440 (2011).
André, P. et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76, 6919–6928 (2002).
Merz, A. et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286, 3018–3032 (2011).
Lindenbach, B.D. et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103, 3805–3809 (2006).
Chang, K.S., Jiang, J., Cai, Z. & Luo, G. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81, 13783–13793 (2007).
Cheng, G. et al. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 105, 3088–3093 (2008).
Burton, D.R., Poignard, P., Stanfield, R.L. & Wilson, I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 (2012).
Gottwein, J.M. et al. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: Role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49, 364–377 (2009).
Meuleman, P. et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology 53, 755–762 (2011).
von Hahn, T. et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132, 667–678 (2007).
Farci, P. et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93, 15394–15399 (1996).
Prentoe, J. et al. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J. Virol. 85, 2224–2234 (2011).
Galun, E. et al. Clinical evaluation (phase I) of a human monoclonal antibody against hepatitis C virus: Safety and antiviral activity. J. Hepatol. 46, 37–44 (2007).
Law, M. et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14, 25–27 (2008).
Giang, E. et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. USA 109, 6205–6210 (2012).
Barth, H. et al. Cellular binding of hepatitis C virus envelope glycoprotein e2 requires cell surface heparan sulfate. J. Biol. Chem. 278, 41003–41012 (2003).
Agnello, V., Abel, G., Elfahal, M., Knight, G.B. & Zhang, Q.X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96, 12766–12771 (1999).
Scarselli, E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 (2002).
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998).
Evans, M.J. et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 (2007).
Ploss, A. et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882–886 (2009).
Shulla, A. & Randall, G. Hepatitis C virus-host interactions, replication, and viral assembly. Curr. Opin. Virol. 2, 725–732 (2012).
Zheng, A. et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 81, 12465–12471 (2007).
Lupberger, J. et al. EGFR and EPHA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17, 589–595 (2011).
Sainz, B. Jr. et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 18, 281–285 (2012).
Blanchard, E. et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80, 6964–6972 (2006).
Tscherne, D.M. et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80, 1734–1741 (2006).
Koutsoudakis, G. et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80, 5308–5320 (2006).
Sharma, N.R. et al. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J. Biol. Chem. 286, 30361–30376 (2011).
Krey, T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762 (2010).
El Omari, K., Iourin, O., Harlos, K., Grimes, J.M. & Stuart, D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 3, 30–35 (2013).
Li, Y., Wang, J., Kanai, R. & Modis, Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. USA 110, 6805–6810 (2013).
Timpe, J.M. et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47, 17–24 (2008).
Meuleman, P. et al. Anti-cd81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48, 1761–1768 (2008).
Meuleman, P. et al. A human monoclonal antibody targeting scavenger receptor class B type I precludes hepatitis C virus infection and viral spread in vitro and in vivo. Hepatology 55, 364–372 (2012).
Lemon, S.M. et al. Development of novel therapies for hepatitis C. Antiviral Res. 86, 79–92 (2010).
Syder, A.J. et al. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J. Hepatol. 54, 48–55 (2011).
Zhu, H. et al. Evaluation of ITC 5061, a scavenger receptor B1 antagonist: Resistance selection and activity in combination with other hepatitis C virus antivirals. J. Infect. Dis. 205, 656–662 (2012).
Liu, R. et al. A peptide derived from hepatitis C virus E2 envelope protein inhibits a post-binding step in HCV entry. Antiviral Res. 86, 172–179 (2010).
Vermehren, J. & Sarrazin, C. The role of resistance in HCV treatment. Best Pract. Res. Clin. Gastroenterol. 26, 487–503 (2012).
Clark, V.C., Peter, J.A. & Nelson, D.R. New therapeutic strategies in HCV: Second-generation protease inhibitors. Liver Int. 33 (suppl. 1), 80–84 (2013).
Lorenz, I.C., Marcotrigiano, J., Dentzer, T.G. & Rice, C.M. Structure of the catalytic domain of the hepatitis C virus NS2–3 protease. Nature 442, 831–835 (2006).
Jirasko, V. et al. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6, e1001233 (2010).
Popescu, C.I. et al. Ns2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7, e1001278 (2011).
Egger, D. et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 (2002).
Romero-Brey, I. et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8, e1003056 (2012).
Lesburg, C.A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6, 937–943 (1999).
Raney, K.D., Sharma, S.D., Moustafa, I.M. & Cameron, C.E. Hepatitis C virus non-structural protein 3 (HCV NS3): A multifunctional antiviral target. J. Biol. Chem. 285, 22725–22731 (2010).
Tellinghuisen, T.L., Marcotrigiano, J., Gorbalenya, A.E. & Rice, C.M. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279, 48576–48587 (2004).
Tellinghuisen, T.L., Marcotrigiano, J. & Rice, C.M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435, 374–379 (2005).
Love, R.A., Brodsky, O., Hickey, M.J., Wells, P.A. & Cronin, C.N. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83, 4395–4403 (2009).
Tellinghuisen, T.L., Foss, K.L., Treadaway, J.C. & Rice, C.M. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82, 1073–1083 (2008).
Appel, N. et al. Essential role of domain III of nonstructural protein 5a for hepatitis C virus infectious particle assembly. PLoS Pathog. 4, e1000035 (2008).
Tellinghuisen, T.L., Foss, K.L. & Treadaway, J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4, e1000032 (2008).
Neddermann, P. et al. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78, 13306–13314 (2004).
Lawitz, E. et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 368, 1878–1887 (2013).
Jacobson, I.M. et al. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N. Engl. J. Med. 368, 1867–1877 (2013).
Arnold, J.J. et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 8, e1003030 (2012).
Gao, M. et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100 (2010).
Scheel, T.K., Gottwein, J.M., Mikkelsen, L.S., Jensen, T.B. & Bukh, J. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not interferon-α. Gastroenterology 140, 1032–1042 (2011).
Guedj, J. et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc. Natl. Acad. Sci. USA 110, 3991–3996 (2013).
Pol, S. et al. Daclatasvir for previously untreated chronic hepatitis C genotype-1 infection: A randomised, parallel-group, double-blind, placebo-controlled, dose-finding, phase 2a trial. Lancet Infect. Dis. 12, 671–677 (2012).
Sulkowski, M. et al. Sustained virologic response with daclatasvir plus sofosbuvir ± ribavirin (RBV) in chronic HCV genotype (GT) 1-infected patients who previously failed telaprevir (TVR) or boceprevir (BOC). J. Hepatol. 58, S570 (2013).
Gu, M. & Rice, C.M. Three conformational snapshots of the hepatitis C virus ns3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. USA 107, 521–528 (2010).
Rai, R. & Deval, J. New opportunities in anti-hepatitis C virus drug discovery: Targeting ns4b. Antiviral Res. 90, 93–101 (2011).
Einav, S. et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat. Biotechnol. 26, 1019–1027 (2008).
Esser-Nobis, K. et al. Analysis of hepatitis C virus resistance to silibinin in vitro and in vivo points to a novel mechanism involving nonstructural protein 4b. Hepatology 57, 953–963 (2013).
Watashi, K., Hijikata, M., Hosaka, M., Yamaji, M. & Shimotohno, K. Cyclosporin a suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38, 1282–1288 (2003).
Paeshuyse, J. et al. The non-immunosuppressive cyclosporin debio-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43, 761–770 (2006).
Hopkins, S. et al. Scy-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob. Agents Chemother. 54, 660–672 (2010).
Yang, F. et al. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82, 5269–5278 (2008).
Rosnoblet, C. et al. Hepatitis C virus NS5B and host cyclophilin a share a common binding site on NS5A. J. Biol. Chem. 287, 44249–44260 (2012).
Hopkins, S. et al. The cyclophilin inhibitor scy-635 suppresses viral replication and induces endogenous interferons in patients with chronic HCV genotype 1 infection. J. Hepatol. 57, 47–54 (2012).
Flisiak, R. et al. The cyclophilin inhibitor debio 025 combined with peg IFN-α2a significantly reduces viral load in treatment-naive hepatitis C patients. Hepatology 49, 1460–1468 (2009).
Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 (2005).
Machlin, E.S., Sarnow, P. & Sagan, S.M. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. USA 108, 3193–3198 (2011).
Jangra, R.K., Yi, M. & Lemon, S.M. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 84, 6615–6625 (2010).
Li, Y., Masaki, T., Yamane, D., McGivern, D.R. & Lemon, S.M. Competing and noncompeting activities of miR-122 and the 5′ exonuclease xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 110, 1881–1886 (2013).
Shimakami, T. et al. Stabilization of hepatitis C virus RNA by an ago2-miR-122 complex. Proc. Natl. Acad. Sci. USA 109, 941–946 (2012).
Lanford, R.E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Li, Y.P., Gottwein, J.M., Scheel, T.K., Jensen, T.B. & Bukh, J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc. Natl. Acad. Sci. USA 108, 4991–4996 (2011).
Hsu, S.H. et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883 (2012).
Tsai, W.C. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122, 2884–2897 (2012).
Janssen, H.L. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013).
Ye, J. et al. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100, 15865–15870 (2003).
Kapadia, S.B. & Chisari, F.V. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 102, 2561–2566 (2005).
Harrison, S.A. et al. Serum cholesterol and statin use predict virological response to peginterferon and ribavirin therapy. Hepatology 52, 864–874 (2010).
Bishé, B., Syed, G. & Siddiqui, A. Phosphoinositides in the hepatitis C virus life cycle. Viruses 4, 2340–2358 (2012).
Berger, K.L. et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III -α in hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 106, 7577–7582 (2009).
Reiss, S. et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9, 32–45 (2011).
Bianco, A. et al. Metabolism of phosphatidylinositol 4-kinase III-α-dependent PI4P is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 8, e1002576 (2012).
Vaillancourt, F.H. et al. Evaluation of phosphatidylinositol-4-kinase III-α as a hepatitis C virus drug target. J. Virol. 86, 11595–11607 (2012).
McLauchlan, J., Lemberg, M.K., Hope, G. & Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21, 3980–3988 (2002).
Boulant, S. et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281, 22236–22247 (2006).
Herker, E. et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 16, 1295–1298 (2010).
Miyanari, Y. et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097 (2007).
Shavinskaya, A., Boulant, S., Penin, F., McLauchlan, J. & Bartenschlager, R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282, 37158–37169 (2007).
Lavie, M., Goffard, A. & Dubuisson, J. Assembly of a functional HCV glycoprotein heterodimer. Curr. Issues Mol. Biol. 9, 71–86 (2007).
Steinmann, E. et al. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 46, 330–338 (2007).
Griffin, S. et al. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology 48, 1779–1790 (2008).
Durantel, D. Celgosivir, an α-glucosidase I inhibitor for the potential treatment of HCV infection. Curr. Opin. Investig. Drugs 10, 860–870 (2009).
Wozniak, A.L. et al. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6, e1001087 (2010).
Tanwandee, T. et al. High sustained viral response with a HCV p7 inhibitor, bit225: Antiviral activity and tolerability of bit225 plus pegylated interferon alfa 2b and weight-based ribavirin for 28 days in HCV treatment-naive patients. Hepatology 56, 1530 (2012).
Huang, H. et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 104, 5848–5853 (2007).
Gastaminza, P. et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82, 2120–2129 (2008).
Counihan, N.A., Rawlinson, S.M. & Lindenbach, B.D. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 7, e1002302 (2011).
Coller, K.E. et al. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 8, e1002466 (2012).
Jiang, J. & Luo, G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83, 12680–12691 (2009).
Hunt, D. & Pockros, P. What are the promising new therapies in the field of chronic hepatitis C after the first-generation direct-acting antivirals? Curr. Gastroenterol. Rep. 15, 303 (2013).
Jacobson, I. et al. Simeprevir (TMC435) with peginterferon/ribavirin for chronic HCV genotype-1 infection in treatment-naïve patients: Results from QUEST-1, a phase III trial. J. Hepatol. 58, S574 (2013).
Manns, M. et al. Simeprevir (TMC435) with peginterferon/ribavirin for treatment of chronic HCV genotype-1 infection in treatment-naïve patients: Results from QUEST-2, a phase III trial. J. Hepatol. 58, S568 (2013).
Ferenci, P. et al. Faldaprevir plus pegylated interferon alfa-2a and ribavirin in chronic HCV genotype-1 treatment-naïve patients: Final results from startverso1, a randomised, double-blind, placebo-controlled phase III trial. J. Hepatol. 58, S569 (2013).
Lok, A.S. et al. Sustained virologic response in chronic HCV genotype (GT) 1-infected null responders with combination of daclatasvir (DCV; NS5A inhibitor) and asunaprevir (ASV; ns3 inhibitor) with or without peginterferon alfa-2a/ribavirin (PEG/RBV). Hepatology 56, 230A–231A (2012).
Thompson, A.J. et al. Six weeks of a NS5A inhibitor (GS-5885), protease inhibitor (GS-9451) plus peginterferon/ribavirin (PR) achieves high SVR4 rates in genotype 1 IL28B CC treatment naive HCV patients: Interim results of a prospective, randomized trial. Hepatology 56, 556A–557A (2012).
Feld, J.J. et al. Up to 100% svr4 rates with ritonavir-boosted danoprevir (DNVR), mericitabine (MCB) and ribavirin (R) ± peginterferon alfa-2a (40kd) (P) in HCV genotype 1-infected partial and null responders: Results from the matterhorn study. Hepatology 56, 231A–232A (2012).
Muir, A.J. et al. Peginterferon lambda-1a (lambda) compared to peginterferon alfa-2a (alfa) in treatment-naive patients with HCV genotypes (GT) 1 or 4: Svr24 results from emerge phase 2b. Hepatology 56, 299A (2012).
Olsen, D.B. et al. Sustained viral response in a hepatitis C virus-infected chimpanzee via a combination of direct-acting antiviral agents. Antimicrob. Agents Chemother. 55, 937–939 (2011).
Ohara, E. et al. Elimination of hepatitis C virus by short term NS3–4A and NS5B inhibitor combination therapy in human hepatocyte chimeric mice. J. Hepatol. 54, 872–878 (2011).
Lok, A.S. et al. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366, 216–224 (2012).
Kowdley, K. et al. Safety and efficacy of interferon-free regimens of ABT-450/R, ABT-267, ABT-333 ± ribavirin in patients with chronic HCV gt1 infection: Results from the aviator study. J. Hepatol. 58, S2 (2013).
Everson, G. et al. Interim analysis of an interferon (IFN)- and ribavirin (RBV)-free regimen of daclatasvir (DCV), asunaprevir (ASV), and BMS-791325 in treatment-naive, hepatitis C virus genotype 1–infected patients. J. Hepatol. 58, S573 (2013).
Gane, E. et al. All-oral sofosbuvir-based 12-week regimens for the treatment of chronic HCV infection: The electron study. J. Hepatol. 58, S6–S7 (2013).
Sulkowski, M. et al. High rate of sustained virologic response with the all-oral combination of daclatasvir (NS5A inhibitor) plus sofosbuvir (nucleotide NS5B inhibitor) with or without ribavirin, in treatment-naive patients chronically infected with HCV GT 1, 2, or 3. Hepatology 56, 1516–1517 (2012).
Poordad, F. et al. Exploratory study of oral combination antiviral therapy for hepatitis C. N. Engl. J. Med. 368, 45–53 (2013).
Jacobson, I.M. et al. VX-222, telaprevir and ribavirin in treatment-naive patients with genotype 1 chronic hepatitis C: Results of the zenith study interferon-free regimen. Hepatology 56, 308A (2012).
Zeuzem, S. et al. An analysis of response rates by fibrosis stage in patients treated with faldaprevir, BI 207127 and ribavirin in the SOUND-C2 study. J. Hepatol. 58, S498 (2013).
Gane, E.J. et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N. Engl. J. Med. 368, 34–44 (2013).
Zeuzem, S. et al. Interferon (IFN)-free combination treatment with the HCV NS3/4A protease inhibitor bi 201335 and the non-nucleoside NS5B inhibitor BI 207127 +/− ribavirin (R): Final results of sound-c2 and predictors of response. Hepatology 56, 308A–309A (2012).
Saeed, M. et al. Efficient replication of genotype 3a and 4a HCV replicons in human hepatoma cells. Antimicrob. Agents Chemother. 56, 5365–5373 (2012).
Li, Y.P. et al. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc. Natl. Acad. Sci. USA 109, 19757–19762 (2012).
Li, Y.P. et al. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc. Natl. Acad. Sci. USA 109, E1101–E1110 (2012).
McPhee, F. et al. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology published online, http://dx.doi.org/10.1002/hep.26388 (2013).
Pawlotsky, J.M. et al. Alisporivir plus ribavirin achieves high rates of sustained HCV clearance (SVR24) as interferon (IFN)-free or IFN-add-on regimen in treatment-naive patients with HCV GT2 or GT3: Final results from vital-1 study. Hepatology 56, 309A–310A (2012).
Marukian, S. et al. Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology 54, 1913–1923 (2011).
Love, R.A. et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87, 331–342 (1996).
Wölk, B. et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74, 2293–2304 (2000).
Moradpour, D., Penin, F. & Rice, C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 (2007).
Kolykhalov, A.A. et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277, 570–574 (1997).
Yanagi, M., Purcell, R.H., Emerson, S.U. & Bukh, J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94, 8738–8743 (1997).
Gottwein, J.M. et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): Genetic analyses and in vivo pathogenesis studies. J. Virol. 84, 5277–5293 (2010).
Bitzegeio, J. et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 6, e1000978 (2010).
Blight, K.J., Kolykhalov, A.A. & Rice, C.M. Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974 (2000).
Bukh, J. et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99, 14416–14421 (2002).
Kaul, A., Woerz, I., Meuleman, P., Leroux-Roels, G. & Bartenschlager, R. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 81, 13168–13179 (2007).
Pietschmann, T. et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103, 7408–7413 (2006).
Imhof, I. & Simmonds, P. Development of an intergenotypic hepatitis C virus (HCV) cell culture method to assess antiviral susceptibilities and resistance development of HCV NS3 protease genes from HCV genotypes 1 to 6. J. Virol. 84, 4597–4610 (2010).
Gastaminza, P., Whitten-Bauer, C. & Chisari, F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA 107, 291–296 (2010).
Chockalingam, K., Simeon, R.L., Rice, C.M. & Chen, Z. A cell protection screen reveals potent inhibitors of multiple stages of the hepatitis C virus life cycle. Proc. Natl. Acad. Sci. USA 107, 3764–3769 (2010).
Acknowledgements
We are grateful to I.M. Jacobson for critical reading of the manuscript. Our research is supported by grants from the US Public Health Service, the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (AI099284, AI072613, AI075099, AI091707, AI090055), Office of the Director through the NIH Roadmap for Medical Research (DK085713), US National Cancer Institute (CA057973), The Rockefeller University Center for Clinical and Translational Science (UL1RR024143), the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust, the Greenberg Medical Research Institute and the Starr Foundation. T.K.H.S. is supported by a postdoctoral fellowship and a Sapere Aude Research Talent Award from The Danish Council for Independent Research. We apologize to the many HCV investigators whose work could not be cited owing to space restrictions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
C.M.R. declares an equity interest in Apath, which holds commercial rights to HCV-related technology, and has in addition served as a consultant or scientific advisor to Genentech, GlaxoSmithKline, iTherX, Merck & Co. Inc. and Novartis. C.M.R. and T.K.H.S. both hold patent rights to HCV-related technology.
Rights and permissions
About this article
Cite this article
Scheel, T., Rice, C. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19, 837–849 (2013). https://doi.org/10.1038/nm.3248
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3248
This article is cited by
-
Mitochondrial DNA copy number in Hepatitis C virus-related chronic liver disease: impact of direct-acting antiviral therapy
Scientific Reports (2023)
-
A single mutation in the E2 glycoprotein of hepatitis C virus broadens the claudin specificity for its infection
Scientific Reports (2022)
-
Computational Design and Analysis of a Poly-Epitope Fusion Protein: A New Vaccine Candidate for Hepatitis and Poliovirus
International Journal of Peptide Research and Therapeutics (2020)
-
Whole-genome sequencing of human Pegivirus variant from an Egyptian patient co-infected with hepatitis C virus: a case report
Virology Journal (2019)
-
Impact of Direct Acting Antivirals on Survival in Patients with Chronic Hepatitis C and Hepatocellular Carcinoma
Scientific Reports (2019)