Abstract
The transcriptional programs that guide lymphocyte differentiation depend on the precise expression and timing of transcription factors (TFs). The TF BACH2 is essential for T and B lymphocytes and is associated with an archetypal super-enhancer (SE). Single-nucleotide variants in the BACH2 locus are associated with several autoimmune diseases, but BACH2 mutations that cause Mendelian monogenic primary immunodeficiency have not previously been identified. Here we describe a syndrome of BACH2-related immunodeficiency and autoimmunity (BRIDA) that results from BACH2 haploinsufficiency. Affected subjects had lymphocyte-maturation defects that caused immunoglobulin deficiency and intestinal inflammation. The mutations disrupted protein stability by interfering with homodimerization or by causing aggregation. We observed analogous lymphocyte defects in Bach2-heterozygous mice. More generally, we observed that genes that cause monogenic haploinsufficient diseases were substantially enriched for TFs and SE architecture. These findings reveal a previously unrecognized feature of SE architecture in Mendelian diseases of immunity: heterozygous mutations in SE-regulated genes identified by whole-exome/genome sequencing may have greater significance than previously recognized.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Bousfiha, A. et al. The 2015 IUIS Phenotypic Classification for Primary Immunodeficiencies. J. Clin. Immunol. 35, 727–738 (2015).
Picard, C. et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J. Clin. Immunol. 35, 696–726 (2015).
Arason, G.J., Jorgensen, G.H. & Ludviksson, B.R. Primary immunodeficiency and autoimmunity: lessons from human diseases. Scand. J. Immunol. 71, 317–328 (2010).
Notarangelo, L.D. Primary immunodeficiencies. J. Allergy Clin. Immunol. 125, S182–S194 (2010).
Conley, M.E. & Casanova, J.-L. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr. Opin. Immunol. 30, 17–23 (2014).
Deau, M.-C. et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J. Clin. Invest. 125, 1764–1765 (2015).
Lo, B. et al. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science 349, 436–440 (2015).
Cunningham-Rundles, C. The many faces of common variable immunodeficiency. Hematology (Am. Soc. Hematol. Educ. Program) 2012, 301–305 (2012).
Rieux-Laucat, F. & Casanova, J.-L. Autoimmunity by haploinsufficiency. Science 345, 1560–1561 (2014).
Lo, B. et al. CHAI and LATAIE: new genetic diseases of CTLA-4 checkpoint insufficiency. Blood 128, 1037–1042 (2016).
Vahedi, G. et al. STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993 (2012).
Whyte, W.A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Roychoudhuri, R. et al. BACH2 represses effector programs to stabilize Treg-mediated immune homeostasis. Nature 498, 506–510 (2013).
Igarashi, K., Ochiai, K., Itoh-Nakadai, A. & Muto, A. Orchestration of plasma cell differentiation by Bach2 and its gene regulatory network. Immunol. Rev. 261, 116–125 (2014).
Ferreira, M.A.R. et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 (2011).
Cooper, J.D. et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40, 1399–1401 (2008).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Dubois, P.C.A. et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 42, 295–302 (2010).
Jin, Y. et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680 (2012).
International Multiple Sclerosis Genetics Consortium. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Nakayama, Y. et al. A limited number of genes are involved in the differentiation of germinal center B cells. J. Cell. Biochem. 99, 1308–1325 (2006).
Ochiai, K. et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J. Biol. Chem. 281, 38226–38234 (2006).
Muto, A. et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429, 566–571 (2004).
Kuwahara, M. et al. The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat. Commun. 5, 3555 (2014).
Povoleri, G.A.M. et al. Thymic versus induced regulatory T cells—who regulates the regulators? Front. Immunol. 4, 169 (2013).
Rosbrook, G.O., Stead, M.A., Carr, S.B. & Wright, S.C. The structure of the Bach2 POZ-domain dimer reveals an intersubunit disulfide bond. Acta Crystallogr. D Biol. Crystallogr. 68, 26–34 (2012).
Uhlig, H.H. et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 147, 990–1007.e3 (2014).
Deane, S., Selmi, C., Naguwa, S.M., Teuber, S.S. & Gershwin, M.E. Common variable immunodeficiency: etiological and treatment issues. Int. Arch. Allergy Immunol. 150, 311–324 (2009).
Salzer, U. & Grimbacher, B. Monogenetic defects in common variable immunodeficiency: what can we learn about terminal B cell differentiation? Curr. Opin. Rheumatol. 18, 377–382 (2006).
Iwata, M. et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004).
Cassani, B. et al. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 141, 2109–2118 (2011).
Igarashi, K., Ochiai, K. & Muto, A. Architecture and dynamics of the transcription factor network that regulates B-to-plasma cell differentiation. J. Biochem. 141, 783–789 (2007).
Seidman, J.G. & Seidman, C. Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 109, 451–455 (2002).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Qian, J. et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159, 1524–1537 (2014).
Huang, N., Lee, I., Marcotte, E.M. & Hurles, M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Creyghton, M.P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Khan, A. & Zhang, X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 44, D164–D171 (2016).
Parker, S.C.J. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA 110, 17921–17926 (2013).
Roychoudhuri, R. et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016).
Shinnakasu, R. et al. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17, 861–869 (2016).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Khoder, A. et al. Regulatory B cells are enriched within the IgM memory and transitional subsets in healthy donors but are deficient in chronic GVHD. Blood 124, 2034–2045 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Naviaux, R.K., Costanzi, E., Haas, M. & Verma, I.M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70, 5701–5705 (1996).
Wingfield, P.T. et al. Biophysical and functional characterization of full-length, recombinant human tissue inhibitor of metalloproteinases-2 (TIMP-2) produced in Escherichia coli. Comparison of wild type and amino-terminal alanine appended variant with implications for the mechanism of TIMP functions. J. Biol. Chem. 274, 21362–21368 (1999).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 32, 894–899 (2011).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34, E2393–E2402 (2013).
Itan, Y. et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods 13, 109–110 (2016).
Dang, V.T., Kassahn, K.S., Marcos, A.E. & Ragan, M.A. Identification of human haploinsufficient genes and their genomic proximity to segmental duplications. Eur. J. Hum. Genet. 16, 1350–1357 (2008).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Bible, P.W. et al. PAPST, a user friendly and powerful Java platform for ChIP-seq peak co-localization analysis and beyond. PLoS One 10, e0127285 (2015).
Aken, B.L. et al. The Ensembl gene annotation system. Database (Oxford) 2016, baw093 (2016).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Zhang, H.-M. et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40, D144–D149 (2012).
Acknowledgements
We thank all subjects and healthy donors for their support, and we thank H. Matthews and C. Neurwirth for coordinating control blood samples. This research was supported by the Intramural Research Programs of NIAMS, the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, Clinical Center, and National Human Genome Research Institute, National Institutes of Health. This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. This work was supported by the Crohn's & Colitis Foundation of America (A.D.J.L. and H.H.U.), the US National Institutes of Health (grant KHL125593A to M.K.), the Sigrid Juselius and Emil Aaltonen Foundations (both to J.G.), the Wellcome Trust (grant 097261/Z/11/Z to B.A.; grant 105663/Z/14/Z to R.R.), the European Molecular Biology Organization (grant ALTF 11602012 to A.N.H.), a Marie Curie fellowship (FP7-PEOPLE-2012-IEF, proposal 330621, to A.N.H.), the Imperial College National Institute for Health Research (NIHR) Biomedical Research Centre (N.C. and P.K.), the Oxford NIHR Biomedical Research Centre (H.H.U.), the Chelsea & Westminster Hospital Charity (C.O'B.), the UK Biotechnology and Biological Sciences Research Council (BB/N0077941/1 to R.R. and M.F.S.), Cancer Research UK (C52623/A22597 to R.R.), the Westminster Medical School Research Trust (P.K.), the Biotechnology and Biological Sciences Research Council (grant BBS/E/B/000C0407 to M.A.L. and I.V.), the Cambridge Trust (I.V.), the Leona M. and Harry B. Helmsley Charitable Trust and ESPGHAN (H.H.U.), the MRC Clinical Sciences Centre (CSC) (T.J.A.) and the CSC Genomics Core Laboratory, and by MRC transition funding (T.J.A.). We acknowledge the contribution of the BRC Gastrointestinal biobank–Oxford IBD cohort study, which is supported by the NIHR Oxford Biomedical Research Centre. We thank G. Vahedi, E. Mathé, S. Parker, C. Kanellopoulou and S. Muljo for critical reading of the manuscript; J. Kabat for help with confocal image analysis; and S.S. De Ravin and H. Malech for advice on the use of MaxCyte. Molecular graphics and analyses were done with the UCSF Chimera package, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). This study used high-performance computational capabilities of Helix Systems at the NIH (http://helix.nih.gov).
Author information
Authors and Affiliations
Contributions
B.A., J.G. and J.V. designed and performed experiments, analyzed data and wrote the manuscript. C.O'B., I.V., F.P.D., A.K., A.N.H., J. Keith, M.F.S., A.S., R.R., M.A.L., O.K., H.-W.S. and Y.Z. performed experiments and/or analyzed data. I.J.F., W.S., T.J.A., P.K. and N.C. provided patient samples and clinical and scientific input. K.M.-R. coordinated patient samples. Patient sequencing and sequence analysis were carried out by J.V., N.C., T.J.A., D.K., M.M., J.D.H., J.M. and Y.Z. A.V.V., N.R.W., H.H.U. and M.K. provided scientific input. P.T.W., I.W.P. and J. Kaufman provided scientific input, performed protein chemistry experiments and analyzed data. N.P.R. provided murine reagents for these experiments. M.J.L., J.J.O'S., N.C. and A.D.J.L. provided scientific input, supervised the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
H.H.U. declares industrial project collaboration (unrelated to the current study) with Lilly, UCB Pharma and Vertex Pharmaceuticals. Travel support was received from Actelion and MSD.
Integrated supplementary information
Supplementary Figure 1 Sanger-sequencing chromatograms for the two families.
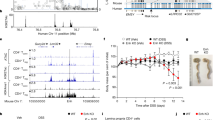
Shown are the Sanger sequencing chromatograms for the two BACH2 mutations (A.II.1 – left; B.II.1 and B.III.2 – right) and unaffected family members (Family A on the left, family B on the right). For each individual the two alleles of the sequenced region and base positions are shown above the sequencing chromatograms. Subject A.II.1 had a heterozygous T to C mutation at coding position 71 whereas patients B.II.1 and B.III.2 were heterozygous for G to A base substitutions at position 2362.
Supplementary Figure 2 Additional phenotyping of cells from affected subjects.
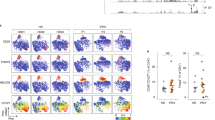
(a) Expression of FOXP3, T-bet, CCR9 and β7-integrin in CD4+ T cells (left four panels, respectively) and frequency of memory B cells (right panel) in peripheral blood of healthy (n=11) and disease controls (n=8), compared to the patients with BACH2 mutations. (b) CD38+CD24+ transitional B cells in patients compared to controls. Shown are representative flow cytometry plots (left) and cumulative data (right) from all patients and matched controls. Patients had elevated proportions of CD38+CD24+ transitional B cells compared to controls but the observed difference does not reach statistical significance. (c) Proliferation (Cell Trace Violet (CTV) dilution) of primary patient CD4+ T cells in response to anti-CD3 and anti-CD28. Shown in b-c are representative FACS plots and cumulative data. Bars show mean ± sem throughout. *p<0.05 **p<0.01 ***p<0.001 ****p<0.0001 by ANOVA (a) and t-test (c).
Supplementary Figure 3 Expression of BACH2 over time, and titration of vector DNA
(a) Immunoblot showing expression of Flag over time in HEK293T cells transfected with Flag-tagged WT and mutant forms of Bach2 (left panel), with Flag quantification (middle panel) and paired Bach2 mRNA expression (right panel) over the same time course. (b) Immunoblot showing expression of Flag 48h after transfection of HEK293T cells with titrated doses of Flag-tagged WT and mutant Bach2-expressing vector (left panel). Flag quantification is shown in the right panel. (a and b) show representative examples of n=2 experiments.
Supplementary Figure 4 Silencing of BACH2 in primary healthy donor cells recapitulates T and B cell phenotypes of affected subjects.
(a and b) BACH2 mRNA expression in primary healthy donor CD4+ T cells (a) and naïve B cells (b) transfected with control RNAi or RNAi specific for BACH2. Shown are mean ± sem from n=6 (a) and n=3 (b) experiments; high (Ramos cell line) and low (Raji cell line) controls for BACH2 expression are additionally shown in a. (c) Proliferation (Cell Trace Violet (CTV) dilution) of primary healthy control T cells transfected with control RNAi or RNAi specific for BACH2 (n=3 from two independent experiments). Shown are representative flow cytometry examples and cumulative mean ± sem from multiple experiments. *p<0.05 **p<0.01 ***p<0.001 by ANOVA.
Supplementary Figure 5 BACH2 coding mutations affect conserved amino acids.
(a) Conservation of both mutated residues across species. (b) Conservation of N-terminus mutation across other BTB/POZ family members. Highlighted in red are the residues mutated in patients, light blue represents non-conserved residues and purple the location of the BTB/POZ domains. Note that the BTB/POZ domain is from residues 27-133 in human BACH2 but that the first α-helix extends from residues 18-34 of the protein. (c) Confocal imaging of HEK293T cells transfected with Flag-tagged murine WT or Bach2E786K and stained for Flag (green) and Hoechst (blue). Insets show enlarged images of single cells and localization of cytoplasmic Flag aggregates (white arrows). Images are representative from 5 independent experiments. The bar graph (right) shows quantification (mean ± sem) of the number of cells containing aggregates per high power field (HPF) from n=5 experiments. White scale bar represents 10 μm in sections and 5 μm in insets. (d) Confocal images of primary lymphocytes from healthy control and patient A.II.1 stained for BACH2 (green) and Hoechst (blue). Scale bars: 5 μm. *p<0.05 by t-test.
Supplementary Figure 6 Mutant forms of Bach2 do not exert dominant negative effects.
(a) Co-immunoprecipitation of Flag-tagged WT with HA-tagged mutant forms of murine Bach2 transfected into HEK293T cells at 1:1 ratio. Shown is a representative example from n=3 independent experiments.
Supplementary Figure 7 Additional phenotyping of T and B cells from Bach2-heterozygous mice.
(a) Expression of Prdm1 mRNA in B cells of Bach2+/+ and Bach2+/– mice. (b) Proportions of CD3+ T cells (top panels), CD4+ and CD8+ T cells (middle panels) and effector CD4+ T cells (CD44+CD62L-; lower panels) in Bach2+/– mice compared to Bach2+/+ littermates. (c) B cell subsets in unimmunized heterozygous mice, showing total B cells (top panels), germinal center (GC) B cells (middle panels) and plasma cells (bottom panels). Data in b and c show representative flow cytometry plots with bar graphs depicting mean ± sem values from a minimum of n=8 mice per group.
Supplementary Figure 8 Super-enhancer (SE) and GWAS association of haploinsufficient genes.
(a) Tissue distribution of genes with SE structure. Shown are only the genes with SE structure in the haplosufficient (HS, top panel), autosomal recessive (AR, middle panel) and haploinsufficient (HI, bottom panel) lists. The presence (red fill) or absence (grey fill) of an SE structure in each tissue is indicated. Each row represents a single gene. Tissue types are shown in columns. Indicated in the “overall” column, is the number of genes with SE architecture in at least one tissue type (see also supplementary Table 3). Source data are indicated. (b-c) Percentage of genes among HS, HI and AR genes that are transcription factors (b) and pie-chart showing function of genes causing HI diseases (c). Source data for b and c are from Qiagen Ingenuity Pathway Analysis. (d) Percentage of SE-regulated genes among transcription factor and non-transcription factor HI genes in comparison to HS and AR genes. (e-g) Ranked order of H3K27Ac-loaded enhancers in human B cells (e), naïve CD8+ (f) and naïve CD4+ (g) T cells. Indicated are the relative positions, ranked according to signal intensity (higher = greater signal intensity), of the top 8 genes from the HI gene list in those cells. Source data from. (h) Percentage of genome-wide association study (GWAS)-associated genes in HS and HI genes compared to all genes, with (right) and without (left) correction for size. Shown are median gene sizes for each gene category. All p-values are from exact Fisher tests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1 and 2, and Supplementary Note.
Supplementary Table 3
Associated super-enhancer (SE) structures in haplosufficient genes and in genes causing haploinsufficiency and autosomal recessive diseases. Gene IDs in each tab are demarcated by tissue as containing (marked as ‘1’) or not containing (marked as ‘0’) an associated SE structure.
Supplementary Table 4
Uniform Resource Locators (URLs) for source data used in Supplementary Table 3.
Supplementary Table 5
Haplosufficient and haploinsufficient genes that have genome-wide association study (GWAS) ‘hits’. Listed are gene identifiers. Please note that BACH2 is included in the haploinsufficient gene list here.
Confocal images from a healthy donor.
Movie from confocal images of lymphocytes of a healthy donor. Green, BACH2; Blue, Hoechst stain.
Confocal images from patient B.II.1.
Movie from confocal images of lymphocytes of BACH2E788K mutant patient. Green, BACH2; Blue, Hoechst stain.
Confocal images from wild-type BACH2 transfected HEK293T cells.
Movies from confocal images of HEK293T cells transfected with Flag-tagged wild-type BACH2. Green, Flag; Blue, Hoechst stain
Confocal images from BACH2E786K-transfected HEK293T cells.
Movies from confocal images of HEK293T cells transfected with Flag-tagged BACH2E786K. Green, Flag; Blue, Hoechst stain.
Rights and permissions
About this article
Cite this article
Afzali, B., Grönholm, J., Vandrovcova, J. et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat Immunol 18, 813–823 (2017). https://doi.org/10.1038/ni.3753
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3753
This article is cited by
-
Tipping the balance in autoimmunity: are regulatory t cells the cause, the cure, or both?
Molecular and Cellular Pediatrics (2024)
-
Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells
Journal of Biomedical Science (2024)
-
Accelerated plasma-cell differentiation in Bach2-deficient mouse B cells is caused by altered IRF4 functions
The EMBO Journal (2024)
-
The role of transcription factors in shaping regulatory T cell identity
Nature Reviews Immunology (2023)
-
An early-onset SLE patient with a novel paternal inherited BACH2 mutation
Journal of Clinical Immunology (2023)