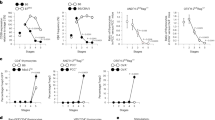
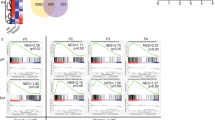
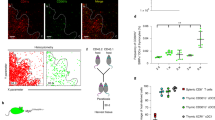
Abstract
Immature CD4+CD8+ (double-positive (DP)) thymocytes are signaled via T cell antigen receptors (TCRs) to undergo positive selection and become responsive to intrathymic cytokines such as interleukin 7 (IL-7). We report here that cytokine signaling is required for positively selected thymocytes to express the transcription factor Runx3, specify CD8 lineage choice and differentiate into cytotoxic-lineage T cells. In DP thymocytes genetically engineered to be cytokine responsive, IL-7 signaling induced TCR-unsignaled DP thymocytes to express Runx3 and to differentiate into mature CD8+ T cells, completely circumventing positive selection. We conclude that TCR-mediated positive selection converts DP cells into cytokine-responsive thymocytes, but it is subsequent signaling by intrathymic cytokines that specifies CD8 lineage choice and promotes differentiation into cytotoxic-lineage T cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jameson, S.C., Hogquist, K.A. & Bevan, M.J. Positive selection of thymocytes. Annu. Rev. Immunol. 13, 93–126 (1995).
Singer, A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol. 14, 207–215 (2002).
Surh, C.D. & Sprent, J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 372, 100–103 (1994).
Van De Wiele, C.J. et al. Thymocytes between the β-selection and positive selection checkpoints are nonresponsive to IL-7 as assessed by STAT-5 phosphorylation. J. Immunol. 172, 4235–4244 (2004).
Yu, Q. et al. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J. Exp. Med. 203, 165–175 (2006).
Chong, M.M. et al. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity 18, 475–487 (2003).
Zamisch, M. et al. Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J. Immunol. 174, 60–67 (2005).
Alves, N.L. et al. Characterization of the thymic IL-7 niche in vivo. Proc. Natl. Acad. Sci. USA 106, 1512–1517 (2009).
Repass, J.F. et al. IL7-hCD25 and IL7-Cre BAC transgenic mouse lines: new tools for analysis of IL-7 expressing cells. Genesis 47, 281–287 (2009).
Singer, A., Adoro, S. & Park, J.H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801 (2008).
Takahama, Y., Suzuki, H., Katz, K.S., Grusby, M.J. & Singer, A. Positive selection of CD4+ T cells by TCR ligation without aggregation even in the absence of MHC. Nature 371, 67–70 (1994).
Ohoka, Y. et al. In vitro differentiation and commitment of CD4+ CD8+ thymocytes to the CD4 lineage, without TCR engagement. Int. Immunol. 8, 297–306 (1996).
Brugnera, E. et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13, 59–71 (2000).
Egawa, T., Tillman, R.E., Naoe, Y., Taniuchi, I. & Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007).
He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005).
He, X. et al. CD4–CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008).
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008).
Sun, G. et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6, 373–381 (2005).
Wang, L. et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat. Immunol. 9, 1122–1130 (2008).
Sato, T. et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22, 317–328 (2005).
Egawa, T. & Littman, D.R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9, 1131–1139 (2008).
Egawa, T. & Taniuchi, I. Antagonistic interplay between ThPOK and Runx in lineage choice of thymocytes. Blood Cells Mol. Dis. 43, 27–29 (2009).
Wang, L. & Bosselut, R. CD4–CD8 lineage differentiation: Thpok-ing into the nucleus. J. Immunol. 183, 2903–2910 (2009).
Cui, Y. et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24, 8037–8047 (2004).
Kaplan, M.H., Schindler, U., Smiley, S.T. & Grusby, M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4, 313–319 (1996).
Hanada, T. et al. A mutant form of JAB/SOCS1 augments the cytokine-induced JAK/STAT pathway by accelerating degradation of wild-type JAB/CIS family proteins through the SOCS-box. J. Biol. Chem. 276, 40746–40754 (2001).
Kubo, M., Hanada, T. & Yoshimura, A. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4, 1169–1176 (2003).
Metcalf, D. The SOCS-1 story. Exp. Hematol. 27, 1715–1723 (1999).
Bosselut, R., Guinter, T.I., Sharrow, S.O. & Singer, A. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes 'paradoxically' appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J. Exp. Med. 197, 1709–1719 (2003).
Lundberg, K., Heath, W., Kontgen, F., Carbone, F.R. & Shortman, K. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4−8+TCRhi thymocytes. J. Exp. Med. 181, 1643–1651 (1995).
Merkenschlager, M. et al. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J. Exp. Med. 200, 1437–1444 (2004).
Suzuki, H., Punt, J.A., Granger, L.G. & Singer, A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity 2, 413–425 (1995).
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002).
Yu, Q., Erman, B., Bhandoola, A., Sharrow, S.O. & Singer, A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J. Exp. Med. 197, 475–487 (2003).
Linette, G.P. et al. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity 1, 197–205 (1994).
Vernachio, J., Li, M., Donnenberg, A.D. & Soloski, M.J. Qa-2 expression in the adult murine thymus. A unique marker for a mature thymic subset. J. Immunol. 142, 48–56 (1989).
El Kassar, N. et al. A dose effect of IL-7 on thymocyte development. Blood 104, 1419–1427 (2004).
Ueno, T. et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200, 493–505 (2004).
Kwan, J. & Killeen, N. CCR7 directs the migration of thymocytes into the thymic medulla. J. Immunol. 172, 3999–4007 (2004).
Park, J.H. et al. 'Coreceptor tuning': cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat. Immunol. 8, 1049–1059 (2007).
Zhu, J. et al. Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J. Exp. Med. 192, 1125–1134 (2000).
Ellmeier, W., Sunshine, M.J., Losos, K., Hatam, F. & Littman, D.R. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 (1997).
Muroi, S. et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 9, 1113–1121 (2008).
Jones, M.E. & Zhuang, Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27, 860–870 (2007).
Hosui, A. et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-β and STAT3 activation. J. Exp. Med. 206, 819–831 (2009).
Sentman, C.L., Shutter, J.R., Hockenbery, D., Kanagawa, O. & Korsmeyer, S.J. Bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 67, 879–888 (1991).
Yu, Q., Erman, B., Park, J.H., Feigenbaum, L. & Singer, A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORγt: impact on thymocyte development. J. Exp. Med. 200, 797–803 (2004).
Kaye, J. et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341, 746–749 (1989).
Marine, J.C. et al. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell 98, 609–616 (1999).
Chu, D.H. et al. Pre-T cell receptor signals are responsible for the down-regulation of Syk protein tyrosine kinase expression. J. Immunol. 163, 2610–2620 (1999).
Acknowledgements
We thank R. Bosselut, N. El Kassar, R. Hodes, D. Singer and N. Taylor for critical reading of the manuscript; S. Habu (Tokai University) for mouse Runx3 cDNA; A. Weiss (University of California San Francisco) for fluorescein isothiocyanate–conjugated monoclonal antibody to Syk (5F5); J. Ihle (St. Jude Children's Research Hospital) for Socs1+/−Ifng−/− mice; and S. Sharrow, A. Adams and L. Granger for flow cytometry. Supported by the Intramural Research Program of the US National Institutes of Health, the National Cancer Institute and the Center for Cancer Research.
Author information
Authors and Affiliations
Contributions
J.-H.P. did experiments, analyzed data and contributed to the writing of the manuscript; S.A., T.G., M.C., M.Y.K. and P.J.L. did experiments and analyzed data; B.E. and A.S.A. constructed some of the experimental mice; Y.C., R.E.G., M.K. and L.H. provided experimental mice; L.F. generated transgenic mice; and A.S. conceptualized the research, directed the study, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 1132 kb)
Rights and permissions
About this article
Cite this article
Park, JH., Adoro, S., Guinter, T. et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol 11, 257–264 (2010). https://doi.org/10.1038/ni.1840
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1840
This article is cited by
-
How autoreactive thymocytes differentiate into regulatory versus effector CD4+ T cells after avoiding clonal deletion
Nature Immunology (2023)
-
Clonal dynamics underlying the skewed CD4/CD8 ratio of mouse thymocytes revealed by TCR-independent barcoding
Communications Biology (2022)
-
An autonomous TCR signal-sensing switch influences CD4/CD8 lineage choice in mice
Communications Biology (2022)
-
Loss of Zfp335 triggers cGAS/STING-dependent apoptosis of post-β selection thymocytes
Nature Communications (2022)
-
Protein abundance of the cytokine receptor γc controls the thymic generation of innate-like T cells
Cellular and Molecular Life Sciences (2022)