Abstract
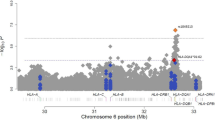
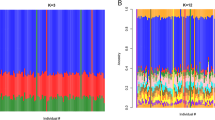
Variants of the gene ALOX5AP (also known as FLAP) encoding arachidonate 5-lipoxygenase activating protein are known to be associated with risk of myocardial infarction1. Here we show that a haplotype (HapK) spanning the LTA4H gene encoding leukotriene A4 hydrolase, a protein in the same biochemical pathway as ALOX5AP, confers modest risk of myocardial infarction in an Icelandic cohort. Measurements of leukotriene B4 (LTB4) production suggest that this risk is mediated through upregulation of the leukotriene pathway. Three cohorts from the United States also show that HapK confers a modest relative risk (1.16) in European Americans, but it confers a threefold larger risk in African Americans. About 27% of the European American controls carried at least one copy of HapK, as compared with only 6% of African American controls. Our analyses indicate that HapK is very rare in Africa and that its occurrence in African Americans is due to European admixture. Interactions with other genetic or environmental risk factors that are more common in African Americans are likely to account for the greater relative risk conferred by HapK in this group.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Helgadottir, A. et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat. Genet. 36, 233–239 (2004).
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 (1959).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Falush, D., Stephens, M. & Pritchard, J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Long, J.C. The genetic structure of admixed populations. Genetics 127, 417–428 (1991).
Parra, E.J. et al. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 63, 1839–1851 (1998).
Adams, J. & Ward, R.H. Admixture studies and the detection of selection. Science 180, 1137–1143 (1973).
Chakraborty, R., Kamboh, M.I., Nwankwo, M. & Ferrell, R.E. Caucasian genes in American blacks: new data. Am. J. Hum. Genet. 50, 145–155 (1992).
McKeigue, P.M., Carpenter, J.R., Parra, E.J. & Shriver, M.D. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann. Hum. Genet. 64, 171–186 (2000).
Reiner, A.P. et al. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am. J. Hum. Genet. 76, 463–477 (2005).
Pritchard, J.K., Stephens, M., Rosenberg, N.A. & Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 67, 170–181 (2000).
Bamshad, M., Wooding, S., Salisbury, B.A. & Stephens, J.C. Deconstructing the relationship between genetics and race. Nat. Rev. Genet. 5, 598–609 (2004).
Jorde, L.B. & Wooding, S.P. Genetic variation, classification and 'race'. Nat. Genet. 36 (Suppl.), S28–S33 (2004).
Royal, C.D. & Dunston, G.M. Changing the paradigm from 'race' to human genome variation. Nat. Genet. 36, S5–S7 (2004).
Tang, H. et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am. J. Hum. Genet. 76, 268–275 (2005).
Barnholtz-Sloan, J.S., Chakraborty, R., Sellers, T.A. & Schwartz, A.G. Examining population stratification via individual ancestry estimates versus self-reported race. Cancer Epidemiol. Biomarkers Prev. 14, 1545–1551 (2005).
Hoggart, C.J. et al. Control of confounding of genetic associations in stratified populations. Am. J. Hum. Genet. 72, 1492–1504 (2003).
Spanbroek, R. et al. Expanding expression of the 5-lipoxygenase pathway within the arterial wall during human atherogenesis. Proc. Natl. Acad. Sci. USA 100, 1238–1243 (2003).
Aiello, R.J. et al. Leukotriene B4 receptor antagonism reduces monocytic foam cells in mice. Arterioscler. Thromb. Vasc. Biol. 22, 443–449 (2002).
Mehrabian, M. et al. Identification of 5-lipoxygenase as a major gene contributing to atherosclerosis susceptibility in mice. Circ. Res. 91, 120–126 (2002).
Samuelsson, B. Leukotrienes: mediators of immediate hypersensitivity reactions and inflammation. Science 220, 568–575 (1983).
Nicholls, S.J. & Hazen, S.L. Myeloperoxidase and cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 25, 1102–1111 (2005).
Brennan, M.L. et al. Prognostic value of myeloperoxidase in patients with chest pain. N. Engl. J. Med. 349, 1595–1604 (2003).
Joint International Society and Federation of Cardiology and World Health Organization Task Force on Standardization of Clinical Nomenclature. Nomenclature and criteria for diagnosis of ischemic heart disease. Circulation 59, 607–609 (1979).
Gulcher, J.R., Kristjansson, K., Gudbjartsson, H. & Stefansson, K. Protection of privacy by third-party encryption in genetic research in Iceland. Eur. J. Hum. Genet. 8, 739–742 (2000).
Gretarsdottir, S. et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 35, 131–138 (2003).
Falk, C.T. & Rubinstein, P. Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann. Hum. Genet. 51, 227–233 (1987).
Terwilliger, J.D. & Ott, J. A haplotype-based 'haplotype relative risk' approach to detecting allelic associations. Hum. Hered. 42, 337–346 (1992).
Smith, M.W. et al. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 74, 1001–1013 (2004).
Acknowledgements
We thank the participants who made this study possible; the nurses at the Icelandic Heart Association; personnel at the deCODE core facilities; A. Rosen, T. Kamineni, J. Pareira and the CRIN staff for recruiting subjects; J. Pritchard for discussion on the admixture analyses; and members of the International HapMap Consortium for providing data which were crucial for our analysis. This work was supported by grants from the US National Institutes of Health (NIH) and by the Emory Alzheimer's Disease Center. Research in Atlanta was supported by NIH grant U54 ES012068 and by Emory General Clinic Research Center grant MO1-RR00039. Research in Cleveland was supported by NIH grant P50 HL077107-01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The following authors are employed by deCODE Genetics and own stock or stock options in the company: A. Helgadottir, A.M., A. Helgason, G.T., U.T., D.F.G., S.G., K.P.M., G.G., A.H., T.J., S.F.A.G., J.S., J.R.G., H.H., A.K. and K.S.
Supplementary information
Supplementary Fig. 1
Genealogical relationship between haplotypes in the LTA4H region. (PDF 812 kb)
Supplementary Fig. 2
Linkage disequilibrium between SNPs in the LTA4H gene region. (PDF 367 kb)
Supplementary Fig. 3
Haplotype risk comparison. (PDF 12 kb)
Supplementary Table 1
SNP allelic assocation to MI. (PDF 77 kb)
Supplementary Table 2
Frequency of haplotypes derived from HapK SNPs. (PDF 91 kb)
Supplementary Table 3
LTB4 levels after ionomycin stimulation of granulocytes. (PDF 57 kb)
Supplementary Table 4
Microsatellite markers used for the evaluation of the European and African ancestry. (PDF 63 kb)
Supplementary Table 5
Markers used to evaluate the local ancestry around LTA4H. (PDF 76 kb)
Rights and permissions
About this article
Cite this article
Helgadottir, A., Manolescu, A., Helgason, A. et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet 38, 68–74 (2006). https://doi.org/10.1038/ng1692
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1692
This article is cited by
-
Contribution of genetic ancestry and polygenic risk score in meeting vitamin B12 needs in healthy Brazilian children and adolescents
Scientific Reports (2021)
-
Impacts of personal DNA ancestry testing
Journal of Community Genetics (2021)
-
In Vivo Cardioprotective Effects and Pharmacokinetic Profile of N-Propyl Caffeamide Against Ischemia Reperfusion Injury
Archivum Immunologiae et Therapiae Experimentalis (2017)
-
Role of methylenetetrahydrofolate reductase 677C→T polymorphism in the development of myocardial infarction: evidence from an original study and updated meta-analysis
Genes & Genomics (2016)
-
Association of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms with myocardial infarction in Tunisian young patients
Comparative Clinical Pathology (2014)