Abstract
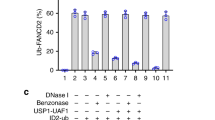
Fanconi anemia is a genetic disease characterized by genomic instability and cancer predisposition1. Nine genes involved in Fanconi anemia have been identified; their products participate in a DNA damage–response network involving BRCA1 and BRCA2 (refs. 2,3). We previously purified a Fanconi anemia core complex containing the FANCL ubiquitin ligase and six other Fanconi anemia–associated proteins4,5,6. Each protein in this complex is essential for monoubiquitination of FANCD2, a key reaction in the Fanconi anemia DNA damage–response pathway2,7. Here we show that another component of this complex, FAAP250, is mutant in individuals with Fanconi anemia of a new complementation group (FA-M). FAAP250 or FANCM has sequence similarity to known DNA-repair proteins, including archaeal Hef, yeast MPH1 and human ERCC4 or XPF. FANCM can dissociate DNA triplex, possibly owing to its ability to translocate on duplex DNA. FANCM is essential for monoubiquitination of FANCD2 and becomes hyperphosphorylated in response to DNA damage. Our data suggest an evolutionary link between Fanconi anemia–associated proteins and DNA repair; FANCM may act as an engine that translocates the Fanconi anemia core complex along DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
BINDPlus
GenBank/EMBL/DDBJ
References
Joenje, H. & Patel, K.J. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2, 446–459 (2001).
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001).
Howlett, N.G. et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297, 606–609 (2002).
Meetei, A.R. et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23, 3417–3426 (2003).
Meetei, A.R. et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35, 165–170 (2003).
Meetei, A.R. et al. X-linked inheritance of Fanconi anemia complementation group B. Nat. Genet. 36, 1219–1224 (2004).
Taniguchi, T. & D'Andrea, A.D. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood 100, 2457–2462 (2002).
Bruun, D. et al. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst.) 2, 1007–1013 (2003).
Komori, K., Fujikane, R., Shinagawa, H. & Ishino, Y. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet. Syst. 77, 227–241 (2002).
Komori, K. et al. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 279, 53175–53185 (2004).
Sgouros, J., Gaillard, P.H. & Wood, R.D. A relationship between a DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem. Sci. 24, 95–97 (1999).
Aravind, L., Walker, D.R. & Koonin, E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242 (1999).
Sijbers, A.M. et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86, 811–822 (1996).
Mu, D., Hsu, D.S. & Sancar, A. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271, 8285–8294 (1996).
Enzlin, J.H. & Scharer, O.D. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053 (2002).
Prakash, R. et al. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 280, 7854–7860 (2005).
Scheller, J., Schurer, A., Rudolph, C., Hettwer, S. & Kramer, W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155, 1069–1081 (2000).
Schurer, K.A., Rudolph, C., Ulrich, H.D. & Kramer, W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics 166, 1673–1686 (2004).
Zavitz, K.H. & Marians, K.J. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J. Biol. Chem. 267, 6933–6940 (1992).
Sung, P., Higgins, D., Prakash, L. & Prakash, S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 7, 3263–3269 (1988).
Van Komen, S., Petukhova, G., Sigurdsson, S., Stratton, S. & Sung, P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6, 563–572 (2000).
Havas, K. et al. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103, 1133–1142 (2000).
Saha, A., Wittmeyer, J. & Cairns, B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16, 2120–2134 (2002).
Levitus, M. et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet., advance online publication 21 August 2005 (10.1038/ng1625).
Cantor, S. et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. USA 101, 2357–2362 (2004).
Pichierri, P. & Rosselli, F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 23, 1178–1187 (2004).
Andreassen, P.R., D'Andrea, A.D. & Taniguchi, T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18, 1958–1963 (2004).
Levitus, M. et al. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood 103, 2498–2503 (2004).
Demuth, I., Digweed, M. & Concannon, P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene 23, 8611–8618 (2004).
Xue, Y. et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 100, 10635–10640 (2003).
Acknowledgements
We thank the individuals with Fanconi anemia and their families for contributing to this study, N. Sherman for mass spectrometry analysis, S. Brill for providing Mus81, R. Wood for XPF, R. Brosh and M. Kenny for RPA, R. Pattni for genotyping, M. Rooimans for technical assistance, the National Cell Culture Center for providing cells and D. Schlessinger for critical reading of the manuscript. Financial support was from the Fanconi anemia Research Fund, Ellison Medical Foundation (W.W.), the US National Institutes of Health (M.H.), Daniel Ayling Fanconi Anaemia Trust (C.G.M.) and the intramural program of National Institute of Health, National Institute on Aging (W.W.). J.P.d.W. and A.L.M. are supported by the Dutch Cancer Society and the Netherlands Organization for Health Research and Development.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
HeLa cells depleted of FAAP250 and FA-M patient cell line deficient of FAAP250 show increased sensitivity to mitomycin C (MMC). (PDF 6 kb)
Supplementary Fig. 2
FANCM (FAAP250) contains a highly-conserved helicase domain. (PDF 352 kb)
Supplementary Fig. 3
The endonuclease domains of the FANCM family of proteins are degenerate at several residues that are conserved in ERCC4/XPF and Hef proteins. (PDF 331 kb)
Supplementary Fig. 4
FANCM shows no detectable helicase activity for several DNA substrates and in the presence of replication protein A (RPA). (PDF 791 kb)
Supplementary Fig. 5
Identification of the FANCM deletion in EUFA867 patient. (PDF 50 kb)
Rights and permissions
About this article
Cite this article
Meetei, A., Medhurst, A., Ling, C. et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37, 958–963 (2005). https://doi.org/10.1038/ng1626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1626
This article is cited by
-
The emergence of a unified mechanism in the Fanconi anemia pathway
Genome Instability & Disease (2021)
-
DONSON and FANCM associate with different replisomes distinguished by replication timing and chromatin domain
Nature Communications (2020)
-
Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion
Nature Communications (2020)
-
FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops
Nature Communications (2019)
-
FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops
Scientific Reports (2019)