Abstract
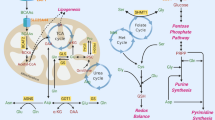
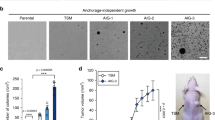
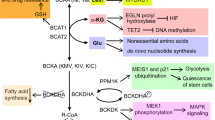
Most tumors exhibit increased glucose metabolism to lactate, however, the extent to which glucose-derived metabolic fluxes are used for alternative processes is poorly understood1,2. Using a metabolomics approach with isotope labeling, we found that in some cancer cells a relatively large amount of glycolytic carbon is diverted into serine and glycine metabolism through phosphoglycerate dehydrogenase (PHGDH). An analysis of human cancers showed that PHGDH is recurrently amplified in a genomic region of focal copy number gain most commonly found in melanoma. Decreasing PHGDH expression impaired proliferation in amplified cell lines. Increased expression was also associated with breast cancer subtypes, and ectopic expression of PHGDH in mammary epithelial cells disrupted acinar morphogenesis and induced other phenotypic alterations that may predispose cells to transformation. Our findings show that the diversion of glycolytic flux into a specific alternate pathway can be selected during tumor development and may contribute to the pathogenesis of human cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vander Heiden, M.G., Cantley, L.C. & Thompson, C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
DeBerardinis, R.J., Lum, J.J., Hatzivassiliou, G. & Thompson, C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 (2008).
Warburg, O., Posener, K. & Negelein, E. Ueber den Stoffwechsel der Tumoren. Biochem. Z. 152, 319–344 (1924).
Bodenhausen, G. & Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 (1980).
Bismut, H., Caron, M., Coudray-Lucas, C. & Capeau, J. Glucose contribution to nucleic acid base synthesis in proliferating hepatoma cells: a glycine-biosynthesis-mediated pathway. Biochem. J. 308, 761–767 (1995).
Snell, K., Natsumeda, Y. & Weber, G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem. J. 245, 609–612 (1987).
Kit, S. The biosynthesis of free glycine and serine by tumors. Cancer Res. 15, 715–718 (1955).
de Koning, T.J. et al. L-serine in disease and development. Biochem. J. 371, 653–661 (2003).
Achouri, Y., Rider, M.H., Van Schaftingen, E. & Robbi, M. Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem. J. 323, 365–370 (1997).
Lu, W., Bennett, B.D. & Rabinowitz, J.D. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871, 236–242 (2008).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Greshock, J. et al. A comparison of DNA copy number profiling platforms. Cancer Res. 67, 10173–10180 (2007).
Slamon, D.J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987).
Luo, J., Solimini, N.L. & Elledge, S.J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009).
Pollari, S. et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 125, 421–430 (2011).
Foulkes, W.D., Smith, I.E. & Reis-Filho, J. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010).
Debnath, J. & Brugge, J.S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5, 675–688 (2005).
Schafer, Z.T. et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113 (2009).
Tabatabaie, L. et al. Novel mutations in 3-phosphoglycerate dehydrogenase (PHGDH) are distributed throughout the protein and result in altered enzyme kinetics. Hum. Mutat. 30, 749–756 (2009).
Thompson, C.B. Metabolic enzymes as oncogenes or tumor suppressors. N. Engl. J. Med. 360, 813–815 (2009).
Teperino, R., Schoonjans, K. & Auwerx, J. Histone methyl transferases and demethylases: can they link metabolism and transcription? Cell Metab. 12, 321–327 (2010).
Nomura, D.K. et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140, 49–61 (2010).
Hara, K. et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 (1998).
Vander Heiden, M.G. et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499 (2010).
Locasale, J.W. & Cantley, L.C. Altered metabolism in cancer. BMC Biol. 8, 88 (2010).
Eng, C.H., Yu, K., Lucas, J., White, E. & Abraham, R.T. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci. Signal. 3, ra31 (2010).
Antoniewicz, M.R., Kelleher, J.K. & Stephanopoulos, G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem. 79, 7554–7559 (2007).
Fernandez, C.A., Des Rosiers, C., Previs, S.F., David, F. & Brunengraber, H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 (1996).
Richardson, A.L. et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132 (2006).
Hoek, K. et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 64, 5270–5282 (2004).
Rhodes, D.R. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 (2004).
Acknowledgements
Microscopy data for this study were acquired and analyzed in the Nikon Imaging Center at Harvard Medical School. J.W.L. was supported by postdoctoral fellowships from the US National Institutes of Health (NIH) and the American Cancer Society. A.R.G. is a recipient of a National Science Foundation (NSF) Graduate Research Fellowship. L.C.C. and J.S.B. were supported by grants from the NIH and the National Cancer Institute (NCI). M.G.V.H. was supported by grants from the NIH, NCI, Smith Family, Damon Runyon Cancer Research Foundation and the Burroughs Wellcome Fund. We thank N. Vena for technical assistance with the FISH analysis and K. Webster and I. Carrecedo for help with immunohistochemistry. We thank J. Rabinowitz, A. Carrecedo and S.-C. Ng for helpful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
J.W.L., M.G.V.H. and L.C.C. designed the study and wrote the paper. J.W.L., C.A.L., E.M., K.R.M., D.A., H.S., M.G.V.H. and T. Melman carried out experiments. J.W.L. and T. Melman carried out computational analyses. A.J.B., R.B. and M.M. provided help with copy number data. L.C. and A.L.R. provided human cancer samples. N.I.V. and A.H.L. carried out the FISH analysis. J.W.L. and J.M.A. carried out the LC/MS/MS experiments. J.W.L., G.H. and G.W. carried out the NMR experiments. J.W.L., N.I.V., C.M.M. and G.S. carried out the GC/MS experiments. M.S. and A.T.S. generated reagents. J.S.B., T. Muranen and A.R.G. carried out experiments involving acinar morphogenesis and imaging analysis.
Corresponding authors
Ethics declarations
Competing interests
J.W.L., M.V.H. and L.C.C. are consultants, scientific advisors and part owners of Agios Pharmaceuticals and hold patents pertaining to targeting cellular metabolism for cancer treatment. Agios Pharmaceuticals is interested in developing therapeutics that target altered metabolism in cancer.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4. (PDF 1929 kb)
Rights and permissions
About this article
Cite this article
Locasale, J., Grassian, A., Melman, T. et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43, 869–874 (2011). https://doi.org/10.1038/ng.890
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.890
This article is cited by
-
Yap/Taz activity is associated with increased expression of phosphoglycerate dehydrogenase that supports myoblast proliferation
Cell and Tissue Research (2024)
-
Unraveling the role of the mitochondrial one-carbon pathway in undifferentiated thyroid cancer by multi-omics analyses
Nature Communications (2024)
-
Bayesian kinetic modeling for tracer-based metabolomic data
BMC Bioinformatics (2023)
-
AMPK-HIF-1α signaling enhances glucose-derived de novo serine biosynthesis to promote glioblastoma growth
Journal of Experimental & Clinical Cancer Research (2023)
-
PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma
Nature Communications (2023)