Abstract
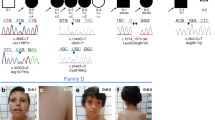
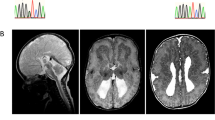
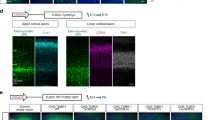
The biological basis for regional and inter-species differences in cerebral cortical morphology is poorly understood. We focused on consanguineous Turkish families with a single affected member with complex bilateral occipital cortical gyration abnormalities. By using whole-exome sequencing, we initially identified a homozygous 2-bp deletion in LAMC3, the laminin γ3 gene, leading to an immediate premature termination codon. In two other affected individuals with nearly identical phenotypes, we identified a homozygous nonsense mutation and a compound heterozygous mutation. In human but not mouse fetal brain, LAMC3 is enriched in postmitotic cortical plate neurons, localizing primarily to the somatodendritic compartment. LAMC3 expression peaks between late gestation and late infancy, paralleling the expression of molecules that are important in dendritogenesis and synapse formation. The discovery of the molecular basis of this unusual occipital malformation furthers our understanding of the complex biology underlying the formation of cortical gyrations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Rakic, P. Specification of cerebral cortical areas. Science 241, 170–176 (1988).
Hofman, M.A. Size and shape of the cerebral cortex in mammals. I. The cortical surface. Brain Behav. Evol. 27, 28–40 (1985).
Caviness, V.S. Jr. Mechanical model of brain convolutional development. Science 189, 18–21 (1975).
Van Essen, D.C. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature 385, 313–318 (1997).
Kriegstein, A., Noctor, S. & Martinez-Cerdeno, V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 7, 883–890 (2006).
Piao, X. et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 303, 2033–2036 (2004).
Kostovic, I. & Rakic, P. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J. Comp. Neurol. 297, 441–470 (1990).
Ferrie, C.D., Jackson, G.D., Giannakodimos, S. & Panayiotopoulos, C.P. Posterior agyria-pachygyria with polymicrogyria: evidence for an inherited neuronal migration disorder. Neurology 45, 150–153 (1995).
Ben Cheikh, B.O. et al. A locus for bilateral occipital polymicrogyria maps to chromosome 6q16-q22. Neurogenetics 10, 35–42 (2009).
Barkovich, A.J., Kuzniecky, R.I., Jackson, G.D., Guerrini, R. & Dobyns, W.B. A developmental and genetic classification for malformations of cortical development. Neurology 65, 1873–1887 (2005).
Bilgüvar, K. et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 (2010).
Sereno, M.I. et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science 268, 889–893 (1995).
Wandell, B.A., Dumoulin, S.O. & Brewer, A.A. Visual field maps in human cortex. Neuron 56, 366–383 (2007).
Dénes, V. et al. Laminin deficits induce alterations in the development of dopaminergic neurons in the mouse retina. Vis. Neurosci. 24, 549–562 (2007).
Johnson, M.B. et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009).
Mrzljak, L., Uylings, H.B., Kostovic, I. & van Eden, C.G. Prenatal development of neurons in the human prefrontal cortex. II. A quantitative Golgi study. J. Comp. Neurol. 316, 485–496 (1992).
Petanjek, Z., Judas, M., Kostovic, I. & Uylings, H.B. Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb. Cortex 18, 915–929 (2008).
Huttenlocher, P.R. & Dabholkar, A.S. Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol. 387, 167–178 (1997).
Durbeej, M. Laminins. Cell Tissue Res. 339, 259–268 (2010).
Helbling-Leclerc, A. et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat. Genet. 11, 216–218 (1995).
Jones, K.J. et al. The expanding phenotype of laminin alpha2 chain (merosin) abnormalities: case series and review. J. Med. Genet. 38, 649–657 (2001).
Gersdorff, N., Kohfeldt, E., Sasaki, T., Timpl, R. & Miosge, N. Laminin γ3 chain binds to nidogen and is located in murine basement membranes. J. Biol. Chem. 280, 22146–22153 (2005).
Koch, M. et al. Characterization and expression of the laminin γ3 chain: a novel, non-basement membrane-associated, laminin chain. J. Cell Biol. 145, 605–618 (1999).
Libby, R.T. et al. Laminin expression in adult and developing retinae: evidence of two novel CNS laminins. J. Neurosci. 20, 6517–6528 (2000).
Yan, H.H. & Cheng, C.Y. Laminin α3 forms a complex with β3 and γ3 chains that serves as the ligand for α 6β1-integrin at the apical ectoplasmic specialization in adult rat testes. J. Biol. Chem. 281, 17286–17303 (2006).
Pinzón-Duarte, G., Daly, G., Li, Y.N., Koch, M. & Brunken, W.J. Defective formation of the inner limiting membrane in laminin α2- and γ3-null mice produces retinal dysplasia. Invest. Ophthalmol. Vis. Sci. 51, 1773–1782 (2010).
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 106, 19096–19101 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Louvi, A., Sisodia, S.S. & Grove, E.A. Presenilin 1 in migration and morphogenesis in the central nervous system. Development 131, 3093–3105 (2004).
Stillman, A.A. et al. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J. Comp. Neurol. 513, 21–37 (2009).
Dennis, G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 (2003).
Engel, S.A., Glover, G.H. & Wandell, B.A. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cereb. Cortex 7, 181–192 (1997).
Song, S.K. et al. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage 17, 1429–1436 (2002).
Jiang, H., van Zijl, P.C., Kim, J., Pearlson, G.D. & Mori, S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Programs Biomed. 81, 106–116 (2006).
Acknowledgements
We are indebted to the subjects and families who have contributed to this study. We would like to thank M. State and J. Noonan for critical comments regarding the study and C. Camputaro for her help with the imaging studies. We acknowledge the use of Yale University Biomedical High Performance Computing Center for data analysis and storage. This study was supported by the Yale Program on Neurogenetics, the Yale Center for Human Genetics and Genomics, and US National Institutes of Health grants RC2NS070477 (to M.G.), UL1RR024139NIH (Yale Clinical and Translational Science Award) and UO1MH081896 (to N.S.). SNP genotyping was supported in part by a US National Institutes of Health Neuroscience Microarray Consortium award U24 NS051869-02S1 (to S.M.).
Author information
Authors and Affiliations
Contributions
M.G. designed the study, and T.B., K.Y.K., A.L., R.P.L., N.S., K.B. and M.G. designed the experiments. T.B., K.Y.K., A.L., K.B., S.Y., M.B., A.O.C., A.K.O. and S.M. performed the experiments. V.D., S.S., B.T., H.K. and C.Y. identified, consented and recruited the study subjects and provided clinical information. A.D. and R.A.B. performed and evaluated magnetic resonance imaging. T.O., H.B., K.D. and E.A. performed and evaluated three-dimensional cortical reconstruction and functional imaging studies. M.C. and R.P.L. developed the bioinformatics scripts for data analysis. W.J.B. provided critical reagents. T.B., T.O., K.Y., K.B., R.P.L. and M.G. analyzed the genetics data. K.Y.K., A.L., Y.Z., N.S. and M.G. analyzed the expression data. T.B., K.Y.K., A.L., R.P.L., N.S., K.B. and M.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors have a provisional patent application under consideration based on the findings of this work.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–7 and Supplementary Figures 1–8. (PDF 1557 kb)
Supplementary Video 1
T2 axial MR images of patient NG 49-1 (MOV 2083 kb)
Supplementary Video 2
T2 sagittal MR images of patient NG 49-1 (MOV 2295 kb)
Supplementary Video 3
Audiovisual EEG monitoring of patient NG 367-1 (MOV 5256 kb)
Supplementary Video 4
T2 axial MR images of patient NG 367-1 (MOV 1410 kb)
Supplementary Video 5
T2 sagittal MR images of patient NG 367-1 (MOV 2570 kb)
Rights and permissions
About this article
Cite this article
Barak, T., Kwan, K., Louvi, A. et al. Recessive LAMC3 mutations cause malformations of occipital cortical development. Nat Genet 43, 590–594 (2011). https://doi.org/10.1038/ng.836
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.836
This article is cited by
-
Mutation of key signaling regulators of cerebrovascular development in vein of Galen malformations
Nature Communications (2023)
-
Deconstructing cortical folding: genetic, cellular and mechanical determinants
Nature Reviews Neuroscience (2019)
-
A Novel S100A8/A9 Induced Fingerprint of Mesenchymal Stem Cells associated with Enhanced Wound Healing
Scientific Reports (2018)
-
A novel mutation in LAMC3 associated with generalized polymicrogyria of the cortex and epilepsy
neurogenetics (2018)
-
Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer
Nature Genetics (2016)