Abstract
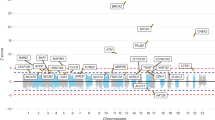
Breast cancer is the most common cancer among women. Common variants at 27 loci have been identified as associated with susceptibility to breast cancer, and these account for ∼9% of the familial risk of the disease. We report here a meta-analysis of 9 genome-wide association studies, including 10,052 breast cancer cases and 12,575 controls of European ancestry, from which we selected 29,807 SNPs for further genotyping. These SNPs were genotyped in 45,290 cases and 41,880 controls of European ancestry from 41 studies in the Breast Cancer Association Consortium (BCAC). The SNPs were genotyped as part of a collaborative genotyping experiment involving four consortia (Collaborative Oncological Gene-environment Study, COGS) and used a custom Illumina iSelect genotyping array, iCOGS, comprising more than 200,000 SNPs. We identified SNPs at 41 new breast cancer susceptibility loci at genome-wide significance (P < 5 × 10−8). Further analyses suggest that more than 1,000 additional loci are involved in breast cancer susceptibility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kamangar, F., Dores, G.M. & Anderson, W.F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 24, 2137–2150 (2006).
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000).
Peto, J. & Mack, T.M. High constant incidence in twins and other relatives of women with breast cancer. Nat. Genet. 26, 411–414 (2000).
Easton, D.F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 (2007).
Hunter, D.J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Stacey, S.N. et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor–positive breast cancer. Nat. Genet. 39, 865–869 (2007).
Stacey, S.N. et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor–positive breast cancer. Nat. Genet. 40, 703–706 (2008).
Ahmed, S. et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 41, 585–590 (2009).
Zheng, W. et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 41, 324–328 (2009).
Thomas, G. et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 41, 579–584 (2009).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Antoniou, A.C. et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor–negative breast cancer in the general population. Nat. Genet. 42, 885–892 (2010).
Fletcher, O. et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl. Cancer Inst. 103, 425–435 (2011).
Haiman, C.A. et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 43, 1210–1214 (2011).
Ghoussaini, M. et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 44, 312–318 (2012).
Siddiq, A. et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21, 5373–5384 (2012).
Eeles, R.A. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. published online; doi:10.1038/ng.2560 (27 March 2013).
Pharoah, P.D.P. et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. published online; doi:10.1038/ng.2564 (27 March 2013).
Couch, F.J. et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 9, e1003212 (2013).
Gaudet, M.M. et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 9, e1003173 (2013).
Cox, A. et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 39, 352–358 (2007).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Lambrechts, D. et al. 11q13 is a susceptibility locus for hormone receptor positive breast cancer. Hum. Mutat. 33, 1123–1132 (2012).
Stevens, K.N. et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 72, 1795–1803 (2012).
Antoniou, A.C. & Easton, D.F. Polygenic inheritance of breast cancer: implications for design of association studies. Genet. Epidemiol. 25, 190–202 (2003).
Figueroa, J.D. et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 20, 4693–4706 (2011).
Mazoyer, S. et al. A polymorphic stop codon in BRCA2. Nat. Genet. 14, 253–254 (1996).
Schutte, M. et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am. J. Hum. Genet. 72, 1023–1028 (2003).
Hemphill, A.W. et al. Mammalian SNM1 is required for genome stability. Mol. Genet. Metab. 94, 38–45 (2008).
Scollen, S. et al. TGF-β signaling pathway and breast cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 20, 1112–1119 (2011).
Ma, X. et al. Pathway analyses identify TGFBR2 as potential breast cancer susceptibility gene: results from a consortium study among Asians. Cancer Epidemiol. Biomarkers Prev. 21, 1176–1184 (2012).
Burwinkel, B. et al. Transcription factor 7–like 2 (TCF7L2) variant is associated with familial breast cancer risk: a case-control study. BMC Cancer 6, 268 (2006).
Goode, E.L. et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 42, 874–879 (2010).
Eeles, R.A. et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 41, 1116–1121 (2009).
Yang, T.P. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26, 2474–2476 (2010).
Shimozawa, N. et al. Identification of a new complementation group of the peroxisome biogenesis disorders and PEX14 as the mutated gene. Hum. Mutat. 23, 552–558 (2004).
Murfuni, I. et al. The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene 32, 610–620 (2013).
Pamidi, A. et al. Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer Res. 67, 8527–8535 (2007).
Leong, S., McKay, M.J., Christopherson, R.I. & Baxter, R.C. Biomarkers of breast cancer apoptosis induced by chemotherapy and TRAIL. J. Proteome Res. 11, 1240–1250 (2012).
Wang, W. et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 173, 395–404 (2006).
Dunbar, M.E., Wysolmerski, J.J. & Broadus, A.E. Parathyroid hormone–related protein: from hypercalcemia of malignancy to developmental regulatory molecule. Am. J. Med. Sci. 312, 287–294 (1996).
Dunbar, M.E. et al. Stromal cells are critical targets in the regulation of mammary ductal morphogenesis by parathyroid hormone–related protein. Dev. Biol. 203, 75–89 (1998).
Qiao, Y. et al. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 71, 3076–3086 (2011).
Kaneda, H. et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 70, 2053–2063 (2010).
Debily, M.A. et al. Expression and molecular characterization of alternative transcripts of the ARHGEF5/TIM oncogene specific for human breast cancer. Hum. Mol. Genet. 13, 323–334 (2004).
Muehlich, S. et al. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene 31, 3913–3923 (2012).
Frayling, T.M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Grant, S.F. et al. Variant of transcription factor 7–like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 38, 320–323 (2006).
Sladek, R. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 (2007).
Jingushi, K. et al. DIF-1 inhibits the Wnt/β-catenin signaling pathway by inhibiting TCF7L2 expression in colon cancer cell lines. Biochem. Pharmacol. 83, 47–56 (2012).
Dantuma, N.P., Heinen, C. & Hoogstraten, D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst.) 8, 449–460 (2009).
Lee, J.C. et al. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochem. Funct. 26, 892–899 (2008).
Castro, P., Liang, H., Liang, J.C. & Nagarajan, L. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics 80, 78–85 (2002).
Sanchez-Cespedes, M. et al. Chromosomal alterations in lung adenocarcinoma from smokers and nonsmokers. Cancer Res. 61, 1309–1313 (2001).
Nakamura, T. et al. Molecular cloning and characterization of Kremen, a novel kringle-containing transmembrane protein. Biochim. Biophys. Acta 1518, 63–72 (2001).
Nakamura, T., Nakamura, T. & Matsumoto, K. The functions and possible significance of Kremen as the gatekeeper of Wnt signalling in development and pathology. J. Cell Mol. Med. 12, 391–408 (2008).
Esseghir, S. et al. Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin. Cancer Res. 13, 3164–3173 (2007).
Morris, A.P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Ahmadiyeh, N. et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA 107, 9742–9746 (2010).
Purcell, S.M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
French, J.D. et al. Functional variants at the 11q13 risk locus regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. published online; 10.1016/j.ajhg.2013.01.002 (27 March 2013).
Bojesen, S.E. et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. published online; doi:10.1038/ng.2566 (27 March 2013).
Aulchenko, Y.S., Struchalin, M.V. & van Duijn, C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Teo, Y.Y. et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 23, 2741–2746 (2007).
Giannoulatou, E., Yau, C., Colella, S., Ragoussis, J. & Holmes, C.C. GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics 24, 2209–2214 (2008).
Haldane, J.B.S. An exact test for randomness of mating. J. Genet. 52, 631–635 (1954).
Stranger, B.E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012).
Acknowledgements
The authors wish to thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. BCAC is funded by Cancer Research UK (C1287/A10118 and C1287/A12014) and by the European Community's Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175) (COGS). Meetings of BCAC have been funded by the European Union European Cooperation in Science and Technology (COST) programme (BM0606). Genotyping of the iCOGS array was funded by the European Union (HEALTH-F2-2009-223175), Cancer Research UK (C1287/A10710), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer program and the Ministry of Economic Development, Innovation and Export Trade of Quebec (grant PSR-SIIRI-701). Combining the GWAS data was supported in part by the US National Institutes of Health (NIH) Cancer Post-Cancer GWAS initiative grant 1 U19 CA 148065-01 (DRIVE, part of the GAME-ON initiative). A full description of funding and acknowledgments is provided in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
K. Michailidou and D.F.E. performed the statistical analysis and drafted the manuscript. D.F.E. conceived and coordinated the synthesis of the iCOGS array and led BCAC. P.H. coordinated COGS. J. Benitez led the iCOGS genotyping working group. A.G.-N., G.P., M.R.A., J. Benitez, D.V., F.B., D.C.T., J. Simard, A.M.D. and C.L. coordinated genotyping of the iCOGS array. M.G.-C., P.D.P.P. and M.K.S. led the BCAC pathology and survival working group. J.C.-C. led the BCAC risk factor working group. A.M.D. and G.C.-T. led the iCOGS quality control working group. J.D., E.D., M. Ghoussaini and A. Lee provided bioinformatics support. M.K.B. and Q. Wang provided data management support for BCAC. S.C. and L.F.A.W. provided analysis of the TCGA expression data. C.T., N.R. and D.F.E. led the UK2 GWAS. O.F., J.P. and I.d.S.S. led the BBCS GWAS. H.N., T.A.M., K. Aittomäki and C.B. led the HEBCS GWAS. P.H., K.C., A.I. and J. Liu led the SASBAC GWAS. Q. Waisfisz, H.M.-H., M.A. and R.B.v.d.L. led the DFBBCS GWAS. J.C.-C., R.H., N.D. and L. Beckman led the MARIE GWAS. A. Meindl, R.K.S., B.M.-M. and P.L. led the GC-HBOC GWAS. J.L.H., M.C.S., E.M., D.F.S. and H.T. led the ABCFS GWAS. A.G.U. and A. Hofman led the genotyping in the Rotterdam study. D.J.H. and S.J.C. led the CGEMS GWAS. F.J.C. and S. Slager coordinated TNBCC. C.A.H., B.E.H., F.S. and L.L.M. coordinated MEC. P.D.P.P., D.F.E. and M. Shah coordinated SEARCH. R.L. coordinated EPIC-Norfolk. J. Brown coordinated SIBS. P.H., K.C., N.S., K.H. and J. Li coordinated SASBAC and pKARMA. S.E.B., B.G.N., S.F.N. and H.F. coordinated CGPS. F.J.C., X.W., C.V. and K.N.S. coordinated MCBCS. D.L., M.M., R.P. and M.-R.C. coordinated LMBC. J.C.-C., A.R., S.N. and D.F.-J. coordinated MARIE. N.J., L.G. and Z.A. coordinated BBCS. K. Aaltonen and T.H. coordinated HEBCS. M.K.S., A.B., L.J.V.t.V. and C.E.v.d.S. coordinated ABCS. P.G., T.T., P.L.-P. and F. Menegaux coordinated CECILE. F. Marme, A. Schneeweiss, C. Sohn and B. Burwinkel coordinated BSUCH. R.L.M., A.G.-N., M.P.Z., J.I.A.P. and J. Benitez coordinated CNIO-BCS. A.C., I.W.B., S.S.C. and M.W.R.R. coordinated SBCS. E.J.S., I.T., M.J.K. and N.M. coordinated BIGGS. I.L.A., J.A.K., G.G. and A.M.M. coordinated OFBCR. A. Lindblom and S. Margolin coordinated KARBAC. M.J.H., A. Hollestelle, A.M.W.v.d.O. and A. Jager coordinated RBCS. J.L.H., M.C.S., Q.M.B., J. Stone, G.S.D. and C.A. coordinated ABCFS. J.L.H., M.C.S., G.G.G., G.S. and L. Baglietto coordinated MCCS. P.A.F., L.H., A.B.E. and M.W.B. coordinated BBCC. H. Brenner, H. Müller, V.A. and C. Stegmaier coordinated ESTHER. A. Swerdlow, A.A., N.O., M.J. and M.G.-C. coordinated UKBGS. M.G.-C., J.F., J. Lissowska and L. Brinton coordinated PBCS. M.S.G., F.L., M.D. and J. Simard coordinated MTLGEBCS. R.W., K.P., A.J.-V. and M. Grip coordinated OBCS. H. Brauch, U.H. and T.B. coordinated GENICA. P.R., P.P., S. Manoukian and B. Bonanni coordinated MBCSG. P.D., R.A.E.M.T., C. Seynaeve and C.J.v.A. coordinated ORIGO. A. Jakubowska, J. Lubinski, K.J. and K.D. coordinated SZBCS. A. Mannermaa, V.K., V.-M.K. and J.M.H. coordinated KBCP. N.V.B., N.N.A. and T.D. coordinated HMBCS. V.N.K. coordinated NBCS. H.A.-C. coordinated UCIBCS. A.E.T. coordinated OSU. S.E. coordinated RPCI. F.F. coordinated DEMOKRITOS. D.K., K.-Y.Y. and D.-Y.N. coordinated SEBCS. K. Matsuo, H. Ito, H. Iwata and A. Sueta coordinated HERPACC. A.H.W., C.-C.T., D.V.D.B. and D.O.S. coordinated LAABC. W.Z., X.-O.S., W.L., Y.-T.G. and H.C. coordinated SGBCS. S.H.T., C.H.Y., S.Y.P. and B.K.C. coordinated MYBRCA. M.H., H. Miao, W.Y.L. and J.-H.S. coordinated SGBCC. K. Muir, A. Lophatananon, S.S.-B. and P.S. coordinated ACP. C.-Y.S., C.-N.H., P.-E.W. and S.-L.D. coordinated TWBCS. S. Sangrajrang, V.G., P.B. and J.M. coordinated TBCS. W.J.B., L.B.S., Q.C. and W.Z. coordinated SCCS. W.Z., S.D.-H., M. Shrubsole and J. Long coordinated NBHS. G.C.-T. coordinated the genotyping component of kConFab. All authors provided critical review of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members is provided in the Supplementary Note.
A list of members is provided in the Supplementary Note.
A list of members is provided in the Supplementary Note.
A list of members is provided in the Supplementary Note.
A list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–8 and Supplementary Note (PDF 2660 kb)
Rights and permissions
About this article
Cite this article
Michailidou, K., Hall, P., Gonzalez-Neira, A. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45, 353–361 (2013). https://doi.org/10.1038/ng.2563
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2563
This article is cited by
-
Differentiated genomic footprints suggest isolation and long-distance migration of Hmong-Mien populations
BMC Biology (2024)
-
The impact of circulating protein levels identified by affinity proteomics on short-term, overall breast cancer risk
British Journal of Cancer (2024)
-
Potential interplay between tumor size and vitamin D receptor (VDR) polymorphisms in breast cancer prognosis: a prospective cohort study
Cancer Causes & Control (2024)
-
BGWAS: Bayesian variable selection in linear mixed models with nonlocal priors for genome-wide association studies
BMC Bioinformatics (2023)
-
An apparent quandary: adoption of polygenics and gene panels for personalised breast cancer risk stratification
BJC Reports (2023)