Abstract
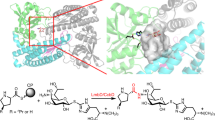
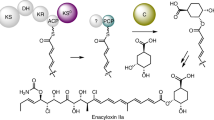
A-503083 B, a capuramycin-type antibiotic, contains an L-aminocaprolactam and an unsaturated hexuronic acid that are linked via an amide bond. A putative class C β-lactamase (CapW) was identified within the biosynthetic gene cluster that—in contrast to the expected β-lactamase activity—catalyzed an amide-ester exchange reaction to eliminate the L-aminocaprolactam with concomitant generation of a small but significant amount of the glyceryl ester derivative of A-503083 B, suggesting a potential role for an ester intermediate in the biosynthesis of capuramycins. A carboxyl methyltransferase, CapS, was subsequently demonstrated to function as an S-adenosylmethionine–dependent carboxyl methyltransferase to form the methyl ester derivative of A-503083 B. In the presence of free L-aminocaprolactam, CapW efficiently converts the methyl ester to A-503083 B, thereby generating a new amide bond. This ATP-independent amide bond formation using methyl esterification followed by an ester-amide exchange reaction represents an alternative to known strategies of amide bond formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Muramatsu, Y. et al. A-503083 A, B, E and F, novel inhibitors of bacterial translocase I, produced by Streptomyces sp. SANK 62799. J. Antibiot. (Tokyo) 57, 639–646 (2004).
Muramatsu, Y. et al. Studies on novel bacterial translocase I inhibitors, A-500359s. I. Taxonomy, fermentation, isolation, physic-chemical properties and structure elucidation of A-500359 A, C, D, and G. J. Antibiot. (Tokyo) 56, 243–252 (2003).
Bugg, T.D.H., Lloyd, A.J. & Roper, D.I. Phospho-MurNAc-pentapeptide translocase (MraY) as a target for antibacterial agents and antibacterial proteins. Infect. Disord. Drug Targets 6, 85–106 (2006).
Suvorov, M., Fisher, J.F. & Mobashery, S. Bacterial cell wall morphology and biochemistry. in Practical Handbook of Microbiology 2nd edn (eds. Goldman, E. & Green, L.H.) 159–190 (CRC Press, Boca Raton, Florida, USA, 2008).
van Heijenoort, J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 71, 620–635 (2007).
Seto, H. et al. The structure of a new nucleoside antibiotic, capuramycin. Tetrahedr. Lett. 29, 2343–2346 (1988).
Funabashi, M. et al. Identification of the biosynthetic gene cluster of A-500359s in Streptomyces griseus SANK60196. J. Antibiot. (Tokyo) 62, 325–332 (2009).
Nolan, E.M. & Walsh, C.T. How nature morphs peptide scaffolds into antibiotics. ChemBioChem 10, 34–53 (2009).
Fisher, J.F., Meroueh, S.O. & Mobashery, S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105, 395–424 (2005).
Lobkovsky, E. et al. Evolution of an enzyme activity: crystallographic structure of 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. USA 90, 11257–11261 (1993).
Santi, D.V., Webster, R.W. Jr. & Cleland, W.W. Kinetics of aminoacyl-tRNA synthetases catalyzed ATP-PPi exchange. Methods Enzymol. 29, 620–627 (1974).
Ohnuki, T., Muramatsu, Y., Miyakoshi, S., Takatsu, T. & Inukai, M. Studies on novel bacterial translocase I inhibitors, A-500359s. IV. Biosynthesis of A-500359s. J. Antibiot. (Tokyo) 56, 268–279 (2003).
Gunstone, F.D. Enzymes as biocatalysts in the modification of natural lipids. J. Sci. Food Agric. 79, 1535–1549 (1999).
Pratt, R.F. Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins). Cell. Mol. Life Sci. 65, 2138–2155 (2008).
Prates, J.A.M. et al. The structure of the feruloyl esterase module of xylanase 10B from the Clostridium thermocellum provides insights into substrate recognition. Structure 9, 1183–1190 (2001).
Matthews, B.W., Sigler, P.B., Henderson, R. & Blow, D.M. Three-dimensional structure of tosyl-α-chymotrypsin. Nature 214, 652–656 (1967).
Lobkovsky, E. et al. Evolution of an enzyme activity: crystallographic structure of 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. USA 90, 11257–11261 (1993).
Awakawa, T. et al. Physically discrete β-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 16, 613–623 (2009).
Schneider, K. et al. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 70, 1417–1423 (2007).
Jensen, S.E. et al. Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 48, 192–202 (2004).
Fischbach, M.A. & Walsh, C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antiobiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Koetsier, M.J., Jekel, P.A., van den Berg, M.A., Bovenberg, R.A. & Janssen, D.B. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem. J. 417, 467–476 (2009).
Tobin, M.B., Fleming, M.D., Skatrud, P.L. & Miller, J.R. Molecular characterization of the acyl-coenzyme A:isopenicillin N acyltransferase gene (penDE) from Penicillium chrysogenum and Aspergillus nidulans and activity of recombinant enzyme in Escherichia coli. J. Bacteriol. 172, 5908–5914 (1990).
Schmutz, E. et al. An unusual amide synthetase (CouL) from the coumermycin A1 biosynthetic gene cluster from Streptomyces rishiriensis DSM 40489. Eur. J. Biochem. 270, 4413–4419 (2003).
Kadi, N., Over-Costales, D., Barona-Gomez, F. & Challis, G.L. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat. Chem. Biol. 3, 652–656 (2007).
Schmelz, S. et al. AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nat. Chem. Biol. 5, 174–182 (2009).
Hollenhorst, M.A., Clardy, J. & Walsh, C.T. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48, 10467–10472 (2009).
Arulanantham, H. et al. ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. J. Biol. Chem. 281, 279–287 (2006).
Gondry, M. et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 5, 414–420 (2009).
Griffith, S.C. et al. Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate. J. Mol. Biol. 313, 1103–1116 (2001).
Cone, M.C., Yin, X., Grochowski, L.L., Parker, M.R. & Zabriskie, T.M. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. ChemBioChem 4, 821–828 (2003).
Palaniappan, N., Ayers, S., Gupta, S., Habib, E.-S. & Reynolds, K.A. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem. Biol. 13, 753–764 (2006).
Saugar, I., Sanz, E., Rubio, M.A., Espinose, J.C. & Jimenez, A. Identification of a set of genes involved in the biosynthesis of the aminonucleoside moiety of antibiotic A201A from Streptomyces capreolus. Eur. J. Biochem. 269, 5527–5535 (2002).
Sugihara, A. et al. A new type of aminoacyltransferase from Saccharothrix sp. AS-2 favorable for the synthesis of d-amino acid-containing peptides. J. Biochem. 131, 247–254 (2002).
Muramatsu, Y. et al. Studies on novel bacterial translocase I inhibitors, A-500359s III. Deaminocaprolactam derivatives of capuramycin: A-500359 E, F, H, M-1 and M-2. J. Antibiot. (Tokyo) 56, 259–267 (2003).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2001).
Kieser, T., Bibb, M., Buttner, M., Chater, K.F. & Hopwood, D.A. Practical Streptomyces Genetics (The John Innes Foundation, Norwich, UK, 2000).
Van Lanen, S.G. et al. Biosynthesis of the β-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring β-amino acyl-S-carrier protein intermediates. J. Am. Chem. Soc. 127, 11594–11595 (2005).
Acknowledgements
This work is supported in part by the Kentucky Science and Technology Corporation (S.V.L.). We thank G. Elliott and J. Jacobsen (Univ. Kentucky) for technical assistance in mass and NMR spectroscopy.
Author information
Authors and Affiliations
Contributions
K.N., M.H., Y.F., T.S. and S.G.V.L. designed the research; M.F., Z.Y., K.N., X.C. and S.G.V.L. performed the experiments; and K.N. and S.G.V.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–11 (PDF 526 kb)
Rights and permissions
About this article
Cite this article
Funabashi, M., Yang, Z., Nonaka, K. et al. An ATP-independent strategy for amide bond formation in antibiotic biosynthesis. Nat Chem Biol 6, 581–586 (2010). https://doi.org/10.1038/nchembio.393
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.393
This article is cited by
-
Actinobacterial chalkophores: the biosynthesis of hazimycins
The Journal of Antibiotics (2024)
-
Mechanism of action of nucleoside antibacterial natural product antibiotics
The Journal of Antibiotics (2019)
-
Peptide Bond Formation via Nα-Protected Diacyldiselenides
International Journal of Peptide Research and Therapeutics (2019)
-
Refining and expanding nonribosomal peptide synthetase function and mechanism
Journal of Industrial Microbiology and Biotechnology (2019)
-
Natural and engineered biosynthesis of nucleoside antibiotics in Actinomycetes
Journal of Industrial Microbiology and Biotechnology (2016)