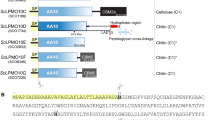
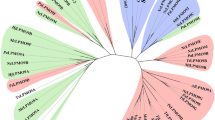
Abstract
Lytic polysaccharide monooxygenases (LPMOs) are a recently discovered class of enzymes capable of oxidizing recalcitrant polysaccharides. They are attracting considerable attention owing to their potential use in biomass conversion, notably in the production of biofuels. Previous studies have identified two discrete sequence-based families of these enzymes termed AA9 (formerly GH61) and AA10 (formerly CBM33). Here, we report the discovery of a third family of LPMOs. Using a chitin-degrading exemplar from Aspergillus oryzae, we show that the three-dimensional structure of the enzyme shares some features of the previous two classes of LPMOs, including a copper active center featuring the 'histidine brace' active site, but is distinct in terms of its active site details and its EPR spectroscopy. The newly characterized AA11 family expands the LPMO clan, potentially broadening both the range of potential substrates and the types of reactive copper-oxygen species formed at the active site of LPMOs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gelfand, I. et al. Sustainable bioenergy production from marginal lands in the US Midwest. Nature 493, 514–517 (2013).
Cantarel, B.L. et al. The Carbohydrate-Active enZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Vaaje-Kolstad, G. et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 (2010).
Phillips, C.M., Beeson, W.T., Cate, J.H. & Marletta, M.A. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 (2011).
Quinlan, R.J. et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 108, 15079–15084 (2011).
Forsberg, Z. et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483 (2011).
Levasseur, A., Drula, E., Lombard, V., Coutinho, P.M. & Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6, 41 (2013).
Karkehabadi, S. et al. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 Å resolution. J. Mol. Biol. 383, 144–154 (2008).
Li, X., Beeson, W.T., Phillips, C.M., Marletta, M.A. & Cate, J.H. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 20, 1051–1061 (2012).
Bey, M. et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl. Environ. Microbiol. 79, 488–496 (2013).
Wu, M. et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the basidiomycota fungus Phanerochaete chrysosporium. J. Biol. Chem. 288, 12828–12839 (2013).
Harris, P.V. et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 (2010).
Hemsworth, G.R., Davies, G.J. & Walton, P.H. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr. Opin. Struct. Biol. 23, 660–668 (2013).
Gilbert, H.J., Knox, J.P. & Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677 (2013).
Boraston, A.B., Bolam, D.N., Gilbert, H.J. & Davies, G.J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004).
Horn, S.J., Vaaje-Kolstad, G., Westereng, B. & Eijsink, V.G.H. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45 (2012).
Petersen, T.N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Flot, J.-F. et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500, 453–457 (2013).
Aachmann, F.L., Sørlie, M., Skjåk-Bræk, G., Eijsink, V.G.H. & Vaaje-Kolstad, G. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl. Acad. Sci. USA 109, 18779–18784 (2012).
Hemsworth, G.R. et al. The copper active site of CBM33 polysaccharide oxygenases. J. Am. Chem. Soc. 135, 6069–6077 (2013).
Wilcox, D.E. Isothermal titration calorimetry of metal ions binding to proteins: An overview of recent studies. Inorg. Chim. Acta 361, 857–867 (2008).
Beeson, W.T., Phillips, C.M., Cate, J.H.D. & Marletta, M.A. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892 (2012).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004).
Kittl, R., Kracher, D., Burgstaller, D., Haltrich, D. & Ludwig, R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 5, 79 (2012).
Peisach, J. & Blumberg, W.E. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch. Biochem. Biophys. 165, 691–708 (1974).
Iwaizumi, M., Kudo, T. & Kita, S. Correlation between the hyperfine coupling constants of donor nitrogens and the structures of the first coordination sphere in copper complexes as studied by nitrogen-14 ENDOR spectroscopy. Inorg. Chem. 25, 1546–1550 (1986).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Acknowledgements
We thank V. Chechik and J. Guo-Wang for assistance with EPR measurements and R. Gregory and J. Robinson for laboratory assistance. This work was funded by the Biotechnology and Biological Sciences Research Council (BB/I014802/1). We thank Diamond Light Source for access to beamlines.
Author information
Authors and Affiliations
Contributions
G.R.H. and P.H.W. performed laboratory experiments; B.H. did the sequence analysis; and G.J.D., B.H., G.R.H. and P.H.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–6 and Supplementary Tables 1–3. (PDF 1580 kb)
Supplementary Data Set 1
Sequences that were retrieved with significant e-values using BAE61530 as the query for a BLAST search against the non-redundant protein sequence database of the NCBI (PDF 484 kb)
Rights and permissions
About this article
Cite this article
Hemsworth, G., Henrissat, B., Davies, G. et al. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol 10, 122–126 (2014). https://doi.org/10.1038/nchembio.1417
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1417
This article is cited by
-
Expanding the catalytic landscape of metalloenzymes with lytic polysaccharide monooxygenases
Nature Reviews Chemistry (2024)
-
Histidine oxidation in lytic polysaccharide monooxygenase
JBIC Journal of Biological Inorganic Chemistry (2023)
-
A pathway for chitin oxidation in marine bacteria
Nature Communications (2022)
-
Quantitative comparison of the biomass-degrading enzyme repertoires of five filamentous fungi
Scientific Reports (2020)
-
Glucose counteracts wood-dependent induction of lignocellulolytic enzyme secretion in monokaryon and dikaryon submerged cultures of the white-rot basidiomycete Pleurotus ostreatus
Scientific Reports (2020)