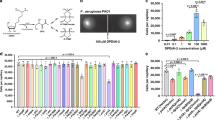
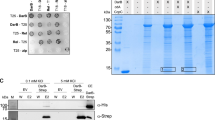
Abstract
Bacteria communicate via small diffusible molecules and thereby mediate group-coordinated behavior, a process referred to as quorum sensing. The prototypical quorum sensing system found in Gram-negative bacteria consists of a LuxI-type autoinducer synthase that produces N-acyl homoserine lactones (AHLs) as signals and a LuxR-type receptor that detects the AHLs to control expression of specific genes. However, many proteobacteria have proteins with homology to LuxR receptors yet lack any cognate LuxI-like AHL synthase. Here we show that in the insect pathogen Photorhabdus luminescens the orphan LuxR-type receptor PluR detects endogenously produced α-pyrones that serve as signaling molecules at low nanomolar concentrations. Additionally, the ketosynthase PpyS was identified as pyrone synthase. Reconstitution of the entire system containing PluR, the PluR-target operon we termed pcf and PpyS in Escherichia coli demonstrated that the cell-cell communication circuit is portable. Our research thus deorphanizes a signaling system and suggests that additional modes of bacterial communication may await discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Waters, C.M. & Bassler, B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005).
Case, R.J., Labbate, M. & Kjelleberg, S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349 (2008).
Marchler-Bauer, A. et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352 (2013).
Fuqua, C. & Greenberg, E.P. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695 (2002).
Fuqua, C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3169–3171 (2006).
Patankar, A.V. & González, J.F. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33, 739–756 (2009).
Subramoni, S. & Venturi, V. LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385 (2009).
Ahmer, B.M.M. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52, 933–945 (2004).
Ferluga, S., Bigirimana, J., Höfte, M. & Venturi, V. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8, 529–538 (2007).
González, J.F. & Venturi, V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 18, 167–174 (2013).
Heermann, R. & Fuchs, T.M. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9, 40 (2008).
Forst, S., Dowds, B., Boemare, N. & Stackebrandt, E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51, 47–72 (1997).
Waterfield, N.R., Ciche, T. & Clarke, D.J. Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 63, 557–574 (2009).
Reimer, D., Pos, K.M., Thines, M., Grün, P. & Bode, H.B. A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat. Chem. Biol. 7, 888–890 (2011).
Chugani, S.A. et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98, 2752–2757 (2001).
Zou, Y. & Nair, S.K. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem. Biol. 16, 961–970 (2009).
Zhang, R.-G. et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417, 971–974 (2002); erratum 476, 240 (2011).
Lintz, M.J., Oinuma, K.-I., Wysoczynski, C.L., Greenberg, E.P. & Churchill, M.E.A. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. USA 108, 15763–15768 (2011).
Erol, O. et al. Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem 11, 1253–1265 (2010).
Joyce, S.A. et al. Bacterial biosynthesis of a multipotent stilbene. Angew. Chem. Int. Edn Engl. 47, 1942–1945 (2008).
Brachmann, A.O. et al. Reciprocal crosstalk between fatty acid and antibiotic biosynthesis in a nematode symbiont. Angew. Chem. Int. Edn. Engl. 51, 12086–12089 (2012).
Goblirsch, B.R., Frias, J.A., Wackett, L.P. & Wilmot, C.M. Crystal structures of Xanthomonas campestris OleA reveal features that promote head-to-head condensation of two long-chain fatty acids. Biochemistry 51, 4138–4146 (2012).
Karlson, P. & Lüscher, M. Pheromones: a new term for a class of biologically active substances. Nature 183, 55–56 (1959).
Duchaud, E. et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–1313 (2003).
Daborn, P.J. et al. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99, 10742–10747 (2002).
Chemler, J.A. et al. Biochemical and structural characterization of germicidin synthase: analysis of a type III polyketide synthase that employs acyl-ACP as a starter unit donor. J. Am. Chem. Soc. 134, 7359–7366 (2012).
Song, L. et al. Type III polyketide synthase β-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J. Am. Chem. Soc. 128, 14754–14755 (2006).
Aoki, Y., Matsumoto, D., Kawaide, H. & Natsume, M. Physiological role of germicidins in spore germination and hyphal elongation in Streptomyces coelicolor A3(2). J. Antibiot. (Tokyo) 64, 607–611 (2011).
Chu, M. et al. Structure of Sch 419560, a novel α-pyrone antibiotic produced by Pseudomonas fluorescens. J. Antibiot. (Tokyo) 55, 215–218 (2002).
Kong, F., Singh, M.P. & Carter, G.T. Pseudopyronines A and B, α-pyrones produced by a marine Pseudomonas sp. F92S91, and evidence for the conversion of 4-hydroxy-α-pyrone to 3-furanone. J. Nat. Prod. 68, 920–923 (2005).
Elbandy, M. et al. α-Pyrones and yellow pigments from the sponge-derived fungus Paecilomyces lilacinus. Bull. Korean Chem. Soc. 30, 188–192 (2009).
Grundmann, F. et al. Identification and isolation of insecticidal oxazoles from Pseudomonas spp. Beilstein J. Org. Chem. 8, 749–752 (2012).
Swem, L.R. et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35, 143–153 (2009).
Bahrani, F.K., Sansonetti, P.J. & Parsot, C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65, 4005–4010 (1997).
Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Weber, K. & Osborn, M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244, 4406–4412 (1969).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular Cloning: a Laboratory Manual (Cold Spring Harbor Laboratory Press, 1989).
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) Method. Methods 25, 402–408 (2001).
Sherlock, O., Schembri, M.A., Reisner, A. & Klemm, P. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186, 8058–8065 (2004).
Bland, J.M. & Altmann, D.G. The logrank test. Br. Med. J. 328, 1073 (2004).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Fechteler, T., Dengler, U. & Schomburg, D. Prediction of protein three-dimensional structures in insertion and deletion regions: a procedure for searching data bases of representative protein fragments using geometric scoring criteria. J. Mol. Biol. 253, 114–131 (1995).
Levitt, M. & Sharon, R. Accurate simulation of protein dynamics in solution. Proc. Natl. Acad. Sci. USA 85, 7557–7561 (1988).
Jones, G., Willett, P., Glen, R.C., Leach, A.R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
Korb, O., Stützle, T. & Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 49, 84–96 (2009).
Corbeil, C.R., Williams, C.I. & Labute, P. Variability in docking success rates due to dataset preparation. J. Comput. Aided Mol. Des. 26, 775–786 (2012).
Acknowledgements
Work in the Heermann lab was financially supported by the Deutsche Forschungsgemeinschaft ((DFG) HE 5247/4-1 and SPP 1617). Work in the Bode lab was supported by the DFG (SPP 1617) and a European Research Council starting grant under grant agreement no. 311477. We thank K. Jung for many helpful scientific discussions; S. Scheu and P. Grün for excellent technical assistance; Q. Zhou for help with HPLC/MS analysis; E.A. Alonso and F.I. Nollmann for help with the PPY production kinetics, S. Kinski for help with ketosynthase mutant construction; and H. Janosz for generation of E. coli insect pathogenicity assays. We are grateful to M. Schobert (Braunschweig) for providing E. coli strain ST18, H. Jung (München) for providing plasmid pUC19-Kan and H. Schweizer (Fort Collins, Colorado) for providing the Tn7 plasmids.
Author information
Authors and Affiliations
Contributions
A.O.B. performed PPY isolation, structure elucidation and quantification and identified ppyS. S.B. performed qRT-PCR, analyzed and quantified PPY bioactivity and specificity for PluR, performed cell clumping assays, generated amino acid replacements in PluR and measured the influence on pyrone sensing. Y.K. constructed PPY-producing E. coli strains. D.K. modeled PluR structure and performed PPY docking and in silico mutagenesis. I.H. performed two-dimensional PAGE, northern blot analyses and Photorhabdus pathogenicity assays. C.M. characterized chemical properties of the PluR signal and performed first E. coli cell clumping assays. K.S. performed preliminary fractionations of P. luminescens supernatants via HPLC and analyzed them for PluR-signal presence. H.B.B. and R.H. coordinated and designed the experiments, analyzed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Results, Supplementary Tables 1-8 and Supplementary Figures 1-17 (PDF 19305 kb)
Rights and permissions
About this article
Cite this article
Brachmann, A., Brameyer, S., Kresovic, D. et al. Pyrones as bacterial signaling molecules. Nat Chem Biol 9, 573–578 (2013). https://doi.org/10.1038/nchembio.1295
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1295
This article is cited by
-
Repurposing host-guest chemistry to sequester virulence and eradicate biofilms in multidrug resistant Pseudomonas aeruginosa and Acinetobacter baumannii
Nature Communications (2023)
-
Exploring AI-2-mediated interspecies communications within rumen microbial communities
Microbiome (2022)
-
The role of bacterial signaling networks in antibiotics response and resistance regulation
Marine Life Science & Technology (2022)
-
Freund oder Feind? — Die zwei Gesichter von Photorhabdus luminescens
BIOspektrum (2021)
-
Symbiosis, virulence and natural-product biosynthesis in entomopathogenic bacteria are regulated by a small RNA
Nature Microbiology (2020)