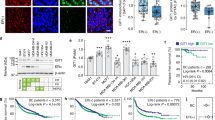
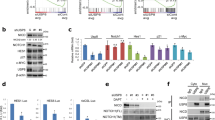
Abstract
Signalling through Notch receptors requires ligand-induced cleavage to release the intracellular domain, which acts as a transcriptional activator in the nucleus. Deregulated Notch1 signalling has been implicated in mammary tumorigenesis; however the mechanisms underlying Notch activation in breast cancer remain unclear. Here, we demonstrate that the prolyl-isomerase Pin1 interacts with Notch1 and affects Notch1 activation. Pin1 potentiates Notch1 cleavage by γ-secretase, leading to an increased release of the active intracellular domain and ultimately enhancing Notch1 transcriptional and tumorigenic activity. We found that Notch1 directly induces transcription of Pin1, thereby generating a positive loop. In human breast cancers, we observed a strong correlation between Pin1 overexpression and high levels of activated Notch1. Thus, the molecular circuitry established by Notch1 and Pin1 may have a key role in cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Radtke, F. & Raj, K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature Rev. Cancer 3, 756–767 (2003).
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell Biol. 7, 678–689 (2006).
Garber, K. Notch emerges as new cancer drug target. J. Natl Cancer Inst. 99, 1284–1285 (2007).
Ayyanan, A. et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc. Natl Acad. Sci. USA 103, 3799–3804 (2006).
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004).
Weijzen, S. et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature Med. 8, 979–986 (2002).
Kiaris, H. et al. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am. J. Pathol. 165, 695–705 (2004).
Stylianou, S., Clarke, R. B. & Brennan, K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517–1525 (2006).
Miele, L., Golde, T. & Osborne, B. Notch signaling in cancer. Curr. Mol. Med. 6, 905–918 (2006).
Yeh, E. S. & Means, A. R. PIN1, the cell cycle and cancer. Nature Rev. Cancer 7, 381–388 (2007).
Lu, K. P. & Zhou, X. Z. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nature Rev. Mol. Cell Biol. 8, 904–916 (2007).
Zacchi, P. et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419, 853–857 (2002).
Mantovani, F. et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nature Struct. Mol. Biol. 14, 912–920 (2007).
Mantovani, F. et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell 14, 625–636 (2004).
Liou, Y. C. et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl Acad. Sci. USA 99, 1335–1340 (2002).
Ryo, A., Nakamura, M., Wulf, G., Liou, Y. C. & Lu, K. P. Pin1 regulates turnover and subcellular localization of β-catenin by inhibiting its interaction with APC. Nature Cell Biol. 3, 793–801 (2001).
Ryo, A. et al. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12, 1413–1426 (2003).
Pastorino, L. et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production. Nature 440, 528–534 (2006).
Akiyama, H., Shin, R. W., Uchida, C., Kitamoto, T. & Uchida, T. Pin1 promotes production of Alzheimer's amyloid β from β-cleaved amyloid precursor protein. Biochem. Biophys. Res. Commun. 336, 521–529 (2005).
Wulf, G., Garg, P., Liou, Y. C., Iglehart, D. & Lu, K. P. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 23, 3397–3407 (2004).
Bao, L. et al. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am. J. Pathol. 164, 1727–1737 (2004).
Wulf, G., Ryo, A., Liou, Y. C. & Lu, K. P. The prolyl isomerase Pin1 in breast development and cancer. Breast Cancer Res. 5, 76–82 (2003).
Jarriault, S. et al. Signalling downstream of activated mammalian Notch. Nature 377, 355–358 (1995).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Zhou, X. Z. et al. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. 6, 873–883 (2000).
Uchida, T. et al. Pin1 and Par14 pe Cell ptidyl prolyl isomerase inhibitors block cell proliferation. Chem. Biol. 10, 15–24 (2003).
Rand, M. D. et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell Biol. 20, 1825–1835 (2000).
Fujimori, F., Takahashi, K., Uchida, C. & Uchida, T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G.(0) arrest. Biochem. Biophys. Res. Commun. 265, 658–663 (1999).
Ingles-Esteve, J., Espinosa, L., Milner, L. A., Caelles, C. & Bigas, A. Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J. Biol. Chem. 276, 44873–44880 (2001).
Ronchini, C. & Capobianco, A. J. Notch(ic)-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene 19, 3914–3924 (2000).
Maier, M. M. & Gessler, M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275, 652–660 (2000).
Osipo, C. et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene (2008).
O'Neill, C. F. et al. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am. J. Pathol. 171, 1023–1036 (2007).
Fortini, M. E. γ-secretase-mediated proteolysis in cell-surface-receptor signalling. Nature Rev. Mol. Cell Biol. 3, 673–684 (2002).
Le Borgne, R., Bardin, A. & Schweisguth, F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762 (2005).
Wolfe, M. S. The γ-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry 45, 7931–7939 (2006).
Struhl, G. & Adachi, A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell 6, 625–636 (2000).
Ramdya, P., Skoch, J., Bacskai, B. J., Hyman, B. T. & Berezovska, O. Activated Notch1 associates with a presenilin-1/γ-secretase docking site. J. Neurochem. 87, 843–850 (2003).
Schroeter, E. H. et al. A presenilin dimer at the core of the γ-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis. Proc. Natl Acad. Sci. USA 100, 13075–13080 (2003).
Kanwar, R. & Fortini, M. E. Notch signaling: a different sort makes the cut. Curr. Biol. 14, R1043–1045 (2004).
Chastagner, P., Israel, A. & Brou, C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3, e2735 (2008).
Takahashi, K. et al. Ablation of a peptidyl prolyl isomerase Pin1 from p53-null mice accelerated thymic hyperplasia by increasing the level of the intracellular form of Notch1. Oncogene 26, 3835–3845 (2007).
Laws, A. M. & Osborne, B. A. p53 regulates thymic Notch1 activation. Eur. J. Immunol. 34, 726–734 (2004).
Rae, J. M., Creighton, C. J., Meck, J M., Haddad, B. R. & Johnson, M. D. MDA-MB-435 cells are derived from M14 melanoma cells — a loss for breast cancer but a boon for melanoma research. Breast Cancer Res. Treat. 104, 13–19 (2007).
Lacroix, M. MDA-MB-435 cells are from melanoma, not breast cancer. Cancer Chemother Pharmacol 63, 567 (2008).
UKCCCR guidelines for the welfare of animals in experimental neoplasia. Cancer Metastasis Rev. 8, 82–88 (1989).
Kononen, J. et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature Med. 4, 844–847 (1998).
Fryer, C. J., White, J. B. & Jones, K. A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520 (2004).
Acknowledgements
We thank LNCIB colleagues, F. Mantovani , L. Collavin and S. Piccolo for critical reading of the manuscript; R. Poddighe for technical support, and M. Stebel and C. Degrassi of the C.S.P.A., University of Trieste. We are grateful to M. Donzelli for generation of the pcDNA3N1ΔE–Flag construct, to T. Sudo for the anti-HES-1 antibody, A. Israel for pGL2-HES-1/LUC, T. Uchida for providing Pin1 knockout mice and to ICGEB for access to their facilities. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC), from Italian University and Research Ministerium (Cofin MIUR), from Friuli-Venezia-Giulia (regional grant), from Association for International Cancer Research (AICR, UK) to G.D.S., and from R01 CA-83736-07 to A.C, AIRC and Fondazione Giancarla Vollaro to S.P. and to Fondazione Monzino and AIRC to P.P.D.F.; M.N. is a FIRC fellow (Fondazione Italiana per la Ricerca sul Cancro). This paper is dedicated to the memory of Stefano Ferrari.
Author information
Authors and Affiliations
Contributions
A.R., L.T., A.S. and M.N. performed biochemical and cell biology experiments; P.N. performed analysis and evaluation of TMA under the supervision of S.P. and P.P.D.F; A.R. performed part of the in vivo experiments; F.K. and A.C. provided essential reagents and assisted with scientific support; G.D.S. was responsible for the overall project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 848 kb)
Rights and permissions
About this article
Cite this article
Rustighi, A., Tiberi, L., Soldano, A. et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol 11, 133–142 (2009). https://doi.org/10.1038/ncb1822
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1822
This article is cited by
-
Expression, Purification, Structural and Functional Characterization of Recombinant Human Parvulin 17
Molecular Biotechnology (2023)
-
Hydroxylation of the NOTCH1 intracellular domain regulates Notch signaling dynamics
Cell Death & Disease (2022)
-
Cancer and Alzheimer’s disease inverse relationship: an age-associated diverging derailment of shared pathways
Molecular Psychiatry (2021)
-
Deficiency of microRNA-628-5p promotes the progression of gastric cancer by upregulating PIN1
Cell Death & Disease (2020)
-
The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers
Scientific Reports (2020)