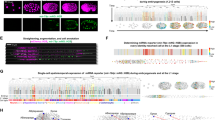
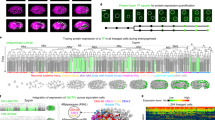
Abstract
Differential regulation of gene expression is essential for cell fate specification in metazoans. Characterizing the transcriptional activity of gene promoters, in time and in space, is therefore a critical step toward understanding complex biological systems. Here we present an in vivo spatiotemporal analysis for ∼900 predicted C. elegans promoters (∼5% of the predicted protein-coding genes), each driving the expression of green fluorescent protein (GFP). Using a flow-cytometer adapted for nematode profiling, we generated 'chronograms', two-dimensional representations of fluorescence intensity along the body axis and throughout development from early larvae to adults. Automated comparison and clustering of the obtained in vivo expression patterns show that genes coexpressed in space and time tend to belong to common functional categories. Moreover, integration of this data set with C. elegans protein-protein interactome data sets enables prediction of anatomical and temporal interaction territories between protein partners.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Davidson, E.H. et al. A genomic regulatory network for development. Science 295, 1669–1678 (2002).
Inoue, T., Wang, M., Ririe, T.O., Fernandes, J.S. & Sternberg, P.W. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. USA 102, 4972–4977 (2005).
Cooper, S.J., Trinklein, N.D., Anton, E.D., Nguyen, L. & Myers, R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 16, 1–10 (2006).
Sulston, J.E., Schierenberg, E., White, J.G. & Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Sulston, J.E. & Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Kimble, J. & Hirsh, D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70, 396–417 (1979).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W. & Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Bao, Z. et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103, 2707–2712 (2006).
The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998).
Vidal, M. A biological atlas of functional maps. Cell 104, 333–339 (2001).
Dupuy, D. et al. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 14, 2169–2175 (2004).
Hobert, O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730 (2002).
McKay, S.J. et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68, 159–169 (2003).
Reece-Hoyes, J.S. et al. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8, 27 (2007).
Mello, C.C., Kramer, J.M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Toms, N., Cooper, J., Patchen, B. & Aamodt, E. High copy arrays containing a sequence upstream of mec-3 alter cell migration and axonal morphology in C. elegans. BMC Dev. Biol. 1, 2 (2001).
Kim, S.K. et al. A gene expression map for Caenorhabditis elegans. Science 293, 2087–2092 (2001).
Li, S. et al. A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 (2004).
Gunsalus, K.C. et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436, 861–865 (2005).
Ge, H., Liu, Z., Church, G.M. & Vidal, M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29, 482–486 (2001).
Rual, J.F. et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Jiang, M. et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98, 218–223 (2001).
Kamath, R.S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003).
Simmer, F. et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1, E12 (2003).
Fernandez, A.G. et al. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 15, 250–259 (2005).
Lundquist, E.A. et al. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122, 1601–1610 (1996).
Spike, C.A., Davies, A.G., Shaw, J.E. & Herman, R.K. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129, 4999–5008 (2002).
Anyanful, A. et al. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J. Cell Biol. 167, 639–647 (2004).
Loria, P.M., Duke, A., Rand, J.B. & Hobert, O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr. Biol. 13, 1317–1323 (2003).
Fujita, M. et al. The role of the ELAV homologue EXC-7 in the development of the Caenorhabditis elegans excretory canals. Dev. Biol. 256, 290–301 (2003).
Han, J.D. et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430, 88–93 (2004).
Chevenet, F., Brun, C., Banuls, A.L., Jacq, B. & Christen, R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439 (2006).
Tavazoie, S., Hughes, J.D., Campbell, M.J., Cho, R.J. & Church, G.M. Systematic determination of genetic network architecture. Nat. Genet. 22, 281–285 (1999).
Thacker, C., Sheps, J.A. & Rose, A.M. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell. Mol. Life Sci. 63, 1193–1204 (2006).
Kim, S. & Wadsworth, W.G. Positioning of longitudinal nerves in C. elegans by nidogen. Science 288, 150–154 (2000).
Sagasti, A., Hobert, O., Troemel, E.R., Ruvkun, G. & Bargmann, C.I. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 13, 1794–1806 (1999).
Kostic, I. & Roy, R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development 129, 2155–2165 (2002).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Harfe, B.D. et al. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12, 2623–2635 (1998).
Hall, D.H. et al. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212, 101–123 (1999).
Bulow, H.E. & Hobert, O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron 41, 723–736 (2004).
Acknowledgements
This work was funded by the National Cancer Institute (NCI 4 R33 CA097516-02)(M.V.), Genome British Columbia, the Canadian Institute of Health Research and Genome Canada (D.B. and D.G.M.), the NetWork Bench National Science Foundation (IIS-0513650), the Muscular Dystrophy Association, the National Institutes of Health (NIH) (1 P20 CA11300-01, A.L.B. and HD43156, W.A.M.) and the Helen Kellogg Institute for International Studies (C.A.H.R.). F.P.R. was supported in part by NIH grant HG003224. M.T. was supported by NIH NRSA Fellowship HG004098. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. Thanks to Tracey Clingingsmith and Abigail Bird for their indispensable assistance and to Michael Cusick and Mike Boxem for careful proofreading of the manuscript.
Author information
Authors and Affiliations
Contributions
Transgenic animals were generated by ballistic transformation by J.S. and J.R.-H. under the supervision of I.A.H., and by microinjection by D.T. and D.L. under the supervision of R.J. and D.B., anatomic annotations of the strains were performed by R.H.-N. and R.V. under the supervision of D.G.M. The chronogram concept was conceived by W.A.M. and implemented by D.D., N.B. and C.A.H. D.D. generated the Gateway promoter::GFP constructs with the help of J.R., and performed the profiling experiments with the technical support of R.P. Computational analyses were performed by N.B., C.A.H., K.V., A.-R.C., A.C. and M.T. under the supervision of D.D., F.P.R., C.L., A.-L.B. and M.V. Lab support was provided by J.R., N.S. and A.B. The manuscript was written by D.D., N.B., C.A.H., A.-L.B. and M.V. The project was conceived and codirected by D.B. and M.V.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Repartition of the transgenic worm strains by origin, grouped by common locus. (PDF 242 kb)
Supplementary Fig. 2
High PCC value between the chronograms correlates with a strong visual resemblance between the fluorescent micrographs of the corresponding animals. (PDF 8567 kb)
Supplementary Fig. 3
Subtrees enrichments. s. (PDF 10891 kb)
Supplementary Fig. 4
Strains BC11521 and BC11545 showed clear expression in the vulval region and the spermatheca, respectively, that was not reported in the initial assessment. (PDF 3070 kb)
Supplementary Table 1
Average images of chronograms grouped in the neighbor joining tree branches that present enrichments in specific GO terms, microarray expression cluster and/or anatomical annotation. (PDF 21900 kb)
Rights and permissions
About this article
Cite this article
Dupuy, D., Bertin, N., Hidalgo, C. et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol 25, 663–668 (2007). https://doi.org/10.1038/nbt1305
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1305
This article is cited by
-
Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma
Journal of Translational Medicine (2018)
-
Dynamic transcriptome profiling towards understanding the morphogenesis and development of diverse feather in domestic duck
BMC Genomics (2018)
-
Combined flow cytometry and high-throughput image analysis for the study of essential genes in Caenorhabditis elegans
BMC Biology (2018)
-
Identification of genes and pathways potentially related to PHF20 by gene expression profile analysis of glioblastoma U87 cell line
Cancer Cell International (2017)
-
Retinoic acid-induced upregulation of miR-219 promotes the differentiation of embryonic stem cells into neural cells
Cell Death & Disease (2017)