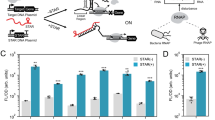
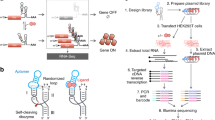
Abstract
Recent studies have demonstrated the importance of noncoding RNA elements in regulating gene expression networks1,2. We describe the design of a class of small trans-acting RNAs that directly regulate gene expression in a ligand-dependent manner. These allosteric riboregulators, which we call antiswitches, are made fully tunable and modular by rational design. They offer flexible control strategies by adopting active or inactive forms in response to ligand binding, depending on their design. They can be tailor-made to regulate the expression of target transcripts in response to different cellular effectors. Coupled with in vitro selection technologies for generating nucleic acid ligand-binding species3,4, antiswitches present a platform for programming cellular behavior and genetic networks with respect to cellular state and environmental stimuli.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Banerjee, D. & Slack, F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays 24, 119–129 (2002).
Kramer, C., Loros, J.J., Dunlap, J.C. & Crosthwaite, S.K. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421, 948–952 (2003).
Ellington, A.D. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Good, L. Diverse antisense mechanisms and applications. Cell. Mol. Life Sci. 60, 823–824 (2003).
Good, L. Translation repression by antisense sequences. Cell. Mol. Life Sci. 60, 854–861 (2003).
Vacek, M., Sazani, P. & Kole, R. Antisense-mediated redirection of mRNA splicing. Cell. Mol. Life Sci. 60, 825–833 (2003).
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Scherer, L. & Rossi, J.J. Recent applications of RNAi in mammalian systems. Curr. Pharm. Biotechnol. 5, 355–360 (2004).
Lilley, D.M. The origins of RNA catalysis in ribozymes. Trends Biochem. Sci. 28, 495–501 (2003).
Mandal, M. & Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 11, 29–35 (2004).
Winkler, W., Nahvi, A. & Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 (2004).
Barrick, J.E. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 101, 6421–6426 (2004).
Yelin, R. et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21, 379–386 (2003).
Lavorgna, G. et al. In search of antisense. Trends Biochem. Sci. 29, 88–94 (2004).
Hermann, T. & Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 287, 820–825 (2000).
Cox, J.C. et al. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res. 30, e108 (2002).
Jhaveri, S., Rajendran, M. & Ellington, A.D. In vitro selection of signaling aptamers. Nat. Biotechnol. 18, 1293–1297 (2000).
Roth, A. & Breaker, R.R. Selection in vitro of allosteric ribozymes. Methods Mol. Biol. 252, 145–164 (2004).
Stojanovic, M.N. & Kolpashchikov, D.M. Modular aptameric sensors. J. Am. Chem. Soc. 126, 9266–9270 (2004).
Smolke, C.D., Carrier, T.A. & Keasling, J.D. Coordinated, differential expression of two genes through directed mRNA cleavage and stabilization by secondary structures. Appl. Environ. Microbiol. 66, 5399–5405 (2000).
Isaacs, F.J. et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22, 841–847 (2004).
Yen, L. et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431, 471–476 (2004).
Buskirk, A.R., Landrigan, A. & Liu, D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 11, 1157–1163 (2004).
Weiss, B., Davidkova, G. & Zhou, L.W. Antisense RNA gene therapy for studying and modulating biological processes. Cell. Mol. Life Sci. 55, 334–358 (1999).
Scherer, L.J. & Rossi, J.J. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 21, 1457–1465 (2003).
Nutiu, R. & Li, Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 125, 4771–4778 (2003).
Zimmermann, G.R., Wick, C.L., Shields, T.P., Jenison, R.D. & Pardi, A. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA 6, 659–667 (2000).
Zimmermann, G.R., Jenison, R.D., Wick, C.L., Simorre, J.P. & Pardi, A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 4, 644–649 (1997).
Gouda, H., Kuntz, I.D., Case, D.A. & Kollman, P.A. Free energy calculations for theophylline binding to an RNA aptamer: Comparison of MM-PBSA and thermodynamic integration methods. Biopolymers 68, 16–34 (2003).
Mathews, D.H. et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 101, 7287–7292 (2004).
Taira, K., Nakagawa, K., Nishikawa, S. & Furukawa, K. Construction of a novel RNA-transcript-trimming plasmid which can be used both in vitro in place of run-off and (G)-free transcriptions and in vivo as multi-sequences transcription vectors. Nucleic Acids Res. 19, 5125–5130 (1991).
Samarsky, D.A. et al. A small nucleolar RNA: ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. USA 96, 6609–6614 (1999).
Mateus, C. & Avery, S.V. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16, 1313–1323 (2000).
Koch, A.L. The metabolism of methylpurines by Escherichia coli. I. Tracer studies. J. Biol. Chem. 219, 181–188 (1956).
Berens, C., Thain, A. & Schroeder, R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 9, 2549–2556 (2001).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Watkins, S.M. & German, J.B. Metabolomics and biochemical profiling in drug discovery and development. Curr. Opin. Mol. Ther. 4, 224–228 (2002).
Khosla, C. & Keasling, J.D. Metabolic engineering for drug discovery and development. Nat. Rev. Drug Discov. 2, 1019–1025 (2003).
Kobayashi, H. et al. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl. Acad. Sci. USA 101, 8414–8419 (2004).
Sambrook, J. & Russell, D.W. Molecular cloning: a laboratory manual, edn. 3 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001).
Sikorski, R.S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989).
Gietz, R. & Woods, R. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. in Guide to Yeast Genetics and Molecular and Cell Biology, Part B, vol. 350 (eds. Guthrie, C. & Fink, G.) 87–96 (Academic Press, San Diego, 2002).
Caponigro, G., Muhlrad, D. & Parker, R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13, 5141–5148 (1993).
Acknowledgements
We thank S.V. Avery, A. Miyawaki and K. Weis for providing genes and plasmids used in assembling the constructs described in this work. This work was supported by startup funds provided by the California Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have a pending patent application whose value may be affected by the publication of this manuscript.
Supplementary information
Supplementary Fig. 1
Sequence and cleavage mechanism of ncRNA expression construct. (PPT 49 kb)
Supplementary Fig. 2
Structural probing of antiswitch s1 through nuclease mapping. (PPT 49 kb)
Supplementary Fig. 3
Sequences and predicted structures of antiswitches s5, s6, and s7 in the absence of ligand binding. (PPT 67 kb)
Supplementary Fig. 4
Sequence and structural switching of a tetracycline-responsive Venus (YFP) regulator, s9, and its target mRNA. (PPT 68 kb)
Supplementary Fig. 5
Set of plasmids used in the in vivo antiswitch characterization studies. (PPT 55 kb)
Supplementary Table 1
Relative RNA levels of target mRNA and antiswitch s1. (PPT 20 kb)
Supplementary Table 2
List of plasmids used in this study. (PPT 39 kb)
Supplementary Table 3
Sequences of antiswitch constructs, controls, and qRT-PCR primers used in this study. (PPT 33 kb)
Rights and permissions
About this article
Cite this article
Bayer, T., Smolke, C. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol 23, 337–343 (2005). https://doi.org/10.1038/nbt1069
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1069
This article is cited by
-
Biomimetic natural biomaterials for tissue engineering and regenerative medicine: new biosynthesis methods, recent advances, and emerging applications
Military Medical Research (2023)
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Nature Communications (2023)
-
Orthogonal control of gene expression in plants using synthetic promoters and CRISPR-based transcription factors
Plant Methods (2022)
-
Engineering synthetic RNA devices for cell control
Nature Reviews Genetics (2022)
-
Enhanced regulation of prokaryotic gene expression by a eukaryotic transcriptional activator
Nature Communications (2021)