Abstract
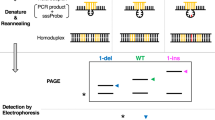
In order to increase the discrimination of single nucleotide polymorphisms in DNA hybridization, artificial mismatches are inserted into probe oligonucleotides using the base analog 3-nitropyrrole. Differences in thermal stability (ΔTm) between hybrids formed with normal and single-nucleotide-variant DNA targets are increased by as much as 200% over conventional hybridization, and are strongly dependent upon the spacing between mismatches. The increased specificity is demonstrated by hybridization analysis and allele-specific amplification within the HLA-DRB locus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Wallace, B.R., Johnson, M.J., Hirose, T., Miyake, T., Kawashima, E.H., and Itakura, K. 1981. The use of synthetic oligonucleotides as hybridization probes. Hybridization of oligonucleotides of mixed sequence to rabbit β-globin DNA. Nucl. Adds Res. 9: 879–894.
Conner, B.J., Reyes, A.A., Morin, C., Itakura, K., Teplitz, R.L., and Wallace, R.B. 1983. Detection of sickle cell βs-globin allele by hybridization with synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 80: 278–282.
Ikuta, S., Takagi, K., Wallace, B.R., and Itakura, K. 1987. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatch base pair. Nucl. Acids Res. 15: 797–811.
Doktycz, M.J., Morris, M.D., Dormady, S.J., Beattie, K.L., and Jacobson, K.B. 1995. Optical melting of 128 octamer DNA duplexes effects of base pair location and nearest neighbors on thermal stability. J. Biol. Chem. 270: 8439–8445.
Southern, E.M., Case-Green, S.C., Elder, J.K., Johnson, M., Mir, K.U., Wang, L., et al. 1994. Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucl. Acids Res. 22: 1368–1373.
Saiki, R.K., Walsh, P.S., Levenson, C.H., and Erlich, H.A. 1989. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 86: 6230–6234.
Breslauer, K.J., Frank, R., Blocker, H., and Marky, L.A. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83: 3746–3750.
McGraw, R.A., Steffe, E.K., and Baxter, S.M. 1990. Sequence-dependent oligonucleotide-target duplex stabilities: rules from empirical studies with a set of twenty-mers. Bio Techniques 8: 674–678.
Tibanyenda, N., De Bruin, S.H., Haasnoot, C.A.G., Van der Marel, G.A., Van Boom, J.H., and Hilbers, C.W., 1984. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G). d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur. J. Biochem. 139: 19–27.
Werntges, H., Steger, G., Riesner, D., and Fritz, H.-J. 1986. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair differencies. Nucl. Acids Res. 14: 3773–3790.
Wenham, P.R., Newton, C.R., and Price, W.H. 1991. Analysis of apolipoprotein E genotypes by the amplification refractory mutation system. Clln. Chem. 37: 241–244.
Newton, C.R., Graham, A., Heptinstall, L.E., Powell, S.J., Summers, C., Kalsheker, N., et al. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucl. Acids Res. 17: 2503–2516.
Ishikawa, Y., Tokunaga, K., Kashiwase, K., Akaza, T., Tadokoro, K. and Juji, T. 1995. Sequence-based typing of HLA-A2 alleles using a primer with an extra base mismatch. Hum. Immunol. 42: 315–318.
Nichols, R., Andrews, P.C., Zhang, P., and Bergstorm, D.E. 1994. A universal nucleoside for use at ambiguous sites in DNA primers. Nature 369: 492–493.
Bergstorm, D.E., Zhang, P., Toma, P.H., Andrews, P.C., and Nichols, R., 1995. Synthesis, structure, and deoxyribonucleoic acid sequencing with a universal nucleoside: 1-(2′-Deoxy-B-D-ribofuranosyl)-3-nitropyrrole. J. Am. Chem. Soc. 117: 1201–1209.
Jacobs, J.W. and Fodor, S.P.A. 1994. Combinational chemistry-applications of light-directed chemical synthesis. TIBTECH 12: 19–26.
Ebel, S., Lane, A.N., and Brawn, T. 1992. Very stable mismatch duplexes: structural and thermodynamic studies on tandem G.A mismatches in DNA. Biochemistry 31: 12083–12086.
Leonard, G.A., Booth, E.D., and Brown, T. 1990. Structural and thermodynamic studies on the adenine/guanine mismatch in B-DNA. Nucl. Acids Res. 18: 5617–5623.
Ke, S.-H. and Wartell, R.M. 1993. Influence of nearest neighbor sequence on the stability of base pair mismatches in long DNA: determination by temperature-gradient gel electrophoresis. Nud. Acids Res. 21: 5137–5143.
Quo, Z., Guilfoyle, R.A., Thiel, A.J., Wang, R., and Smith, L.M. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucl. Adds Res. 22: 5456–5465.
Bein, G., Glaser, R., and Kirchner, H. 1992. Rapid HLA-DRB1 genotyping by nested PCR amplification. Tissue Antigens 39: 68–73.
Olerup, O. and Zetterquist, H. 1992. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 39: 225–235.
Dynabeads M-280 Technical Handbook: Magnetic DNA Technology 6. 1989. Dynal Inc., Great Neck, NY.
Smith, L.M., Kaiser, R.J., Sanders, J.Z., and Hood, L.E. 1987. The synthesis and use of fluorescent oligonucleotides in DNA sequence analysis. Methods Enzymol. 55: 260–301.
Baxter-Lowe, L., Hunter, J., Casper, J., and Gorski, J. 1989. HLA gene amplification and hybridization analysis of polymorphism. HLA matching for bone marrow transplantation of a patent with HLA deficient severe combined Immundeficiency syndrome. J. Clin. Invest. 84: 613–618.
Bodmer, J., Marsh, S., and Albert, E. 1992. Nomenclature for factors of the HLA System. Tissue Antigens 39: 161–173.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Guo, Z., Liu, Q. & Smith, L. Enhanced discrimination of single nucleotide polymorphisms by artificial mismatch hybridization. Nat Biotechnol 15, 331–335 (1997). https://doi.org/10.1038/nbt0497-331
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0497-331
This article is cited by
-
Mismatch-introduced DNA probes constructed on the basis of thermodynamic analysis enable the discrimination of single nucleotide variants
Analytical and Bioanalytical Chemistry (2022)
-
Small molecule electro-optical binding assay using nanopores
Nature Communications (2019)
-
Conditionally fluorescent molecular probes for detecting single base changes in double-stranded DNA
Nature Chemistry (2013)
-
Effect of LNA- and OMeN-modified oligonucleotide probes on the stability and discrimination of mismatched base pairs of duplexes
Journal of Biosciences (2012)
-
IMP PCR primers detect single nucleotide polymorphisms for Anopheles gambiae species identification, Mopti and Savanna rDNA types, and resistance to dieldrin in Anopheles arabiensis
Malaria Journal (2006)