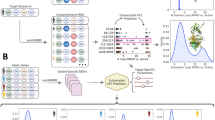
Abstract
We present a technology to screen millions of B cells for natively paired human antibody repertoires. Libraries of natively paired, variable region heavy and light (VH:VL) amplicons are expressed in a yeast display platform that is optimized for human Fab surface expression. Using our method we identify HIV-1 broadly neutralizing antibodies (bNAbs) from an HIV-1 slow progressor and high-affinity neutralizing antibodies against Ebola virus glycoprotein and influenza hemagglutinin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Chan, A.C. & Carter, P.J. Nat. Rev. Immunol. 10, 301–316 (2010).
Brekke, O.H. & Sandlie, I. Nat. Rev. Drug Discov. 2, 52–62 (2003).
Corti, D. & Lanzavecchia, A. Annu. Rev. Immunol. 31, 705–742 (2013).
Burton, D.R. & Hangartner, L. Annu. Rev. Immunol. 34, 635–659 (2016).
Bradbury, A.R.M., Sidhu, S., Dübel, S. & McCafferty, J. Nat. Biotechnol. 29, 245–254 (2011).
Ecker, D.M., Jones, S.D. & Levine, H.L. MAbs 7, 9–14 (2015).
Wilson, P.C. & Andrews, S.F. Nat. Rev. Immunol. 12, 709–719 (2012).
Georgiou, G. et al. Nat. Biotechnol. 32, 158–168 (2014).
Jayaram, N., Bhowmick, P. & Martin, A.C.R. Protein Eng. Des. Sel. 25, 523–529 (2012).
Ponsel, D., Neugebauer, J., Ladetzki-Baehs, K. & Tissot, K. Molecules 16, 3675–3700 (2011).
DeKosky, B.J. et al. Nat. Med. 21, 86–91 (2015).
McDaniel, J.R., DeKosky, B.J., Tanno, H., Ellington, A.D. & Georgiou, G. Nat. Protoc. 11, 429–442 (2016).
Spadiut, O., Capone, S., Krainer, F., Glieder, A. & Herwig, C. Trends Biotechnol. 32, 54–60 (2014).
Bowley, D.R., Labrijn, A.F., Zwick, M.B. & Burton, D.R. Protein Eng. Des. Sel. 20, 81–90 (2007).
Feldhaus, M.J. et al. Nat. Biotechnol. 21, 163–170 (2003).
Lee, J. et al. Nat. Med. 22, 1456–1464 (2016).
Wentz, A.E. & Shusta, E.V. Appl. Environ. Microbiol. 73, 1189–1198 (2007).
Ojima-Kato, T. et al. Protein Eng. Des. Sel. 29, 149–157 (2016).
Kong, R. et al. Science 352, 828–833 (2016).
Stanley, D.A. et al. Nat. Med. 20, 1126–1129 (2014).
Maruyama, T. et al. J. Virol. 73, 6024–6030 (1999).
Tian, M. et al. Cell 166, 1471–1484. e18 (2016).
Joyce, M.G. et al. Cell 166, 609–623 (2016).
Moody, M.A. et al. PLoS One 6, e25797 (2011).
Pinna, D., Corti, D., Jarrossay, D., Sallusto, F. & Lanzavecchia, A. Eur. J. Immunol. 39, 1260–1270 (2009).
Whittle, J.R. et al. J. Virol. 88, 4047–4057 (2014).
Kanekiyo, M. et al. Cell 162, 1090–1100 (2015).
Tillotson, B.J., Cho, Y.K. & Shusta, E.V. Methods 60, 27–37 (2013).
Wang, X.X., Cho, Y.K. & Shusta, E.V. Nat. Methods 4, 143–145 (2007).
Fang, Y., Chu, T.H., Ackerman, M.E. & Griswold, K.E. MAbs 9, 1253–1261 (2017).
Côté, M. et al. Nature 477, 344–348 (2011).
Misasi, J. et al. Science 351, 1343–1346 (2016).
Whittle, J.R.R. et al. Proc. Natl. Acad. Sci. USA 108, 14216–14221 (2011).
Lee, J.E. et al. Nature 454, 177–182 (2008).
Olinger, G.G. Jr. et al. Proc. Natl. Acad. Sci. USA 109, 18030–18035 (2012).
Lavinder, J.J. et al. Proc. Natl. Acad. Sci. USA 111, 2259–2264 (2014).
Doria-Rose, N.A. et al. J. Virol. 83, 188–199 (2009).
Doria-Rose, N.A. et al. J. Virol. 84, 1631–1636 (2010).
Benatuil, L., Perez, J.M., Belk, J. & Hsieh, C.M. Protein Eng. Des. Sel. 23, 155–159 (2010).
Reich, L.L., Dutta, S. & Keating, A.E. J. Mol. Biol. 427, 2135–2150 (2015).
Wang, B. et al. MAbs 8, 1035–1044 (2016).
DeKosky, B.J. et al. Proc. Natl. Acad. Sci. USA 113, E2636–E2645 (2016).
Cale, E.M. et al. Immunity 46, 777–791. e10 (2017).
Whitehead, T.A. et al. Nat. Biotechnol. 30, 543–548 (2012).
DeKosky, B.J. et al. Nat. Biotechnol. 31, 166–169 (2013).
Wang, B. et al. Sci. Rep. 5, 13926 (2015).
Wu, X. et al. Science 329, 856–861 (2010).
Sullivan, N.J. et al. PLoS Med. 3, 0865–0873 (2006).
Yang, Z.Y. et al. J. Virol. 78, 4029–4036 (2004).
Acknowledgements
We gratefully acknowledge B. Hartman for assistance with figures, N. Doria-Rose, G. Ippolito, and M. Kanekiyo for advice and guidance, S. Lucas, and K. Zhou for help with experiments, S. Darko for assistance with data processing, and E. Shusta and S. Harrison for kindly sharing reagents and research tools. This work was funded in part by the intramural research program of the Vaccine Research Center, NIAID, NIH, NIH grant DP5OD023118-01 to B.J.D., NIH grant 5R21CA191239-01 to A.D.E., Leidos Biomedical Research Inc. contract 15X219, DTRA contract HDTRA1-12-C-0105, and NIH grant 1R56AI106006 to G.G.
Author information
Authors and Affiliations
Contributions
B.W., B.J.D., J.R. Mascola, and G.G. conceived the study and designed the experiments. B.W. and B.J.D. conducted the experiments with help from M.R.T., J.L., E.N., J.M., R.K., J.R. McDaniel, G.D., K.E.L., T.N., C.W.C., E.G.V., A.F., A.C., A.P., K.L., E.S.Y., W.-P.K., W.N.V., A.G.S., M.A.M., D.R.A., A.R.H., F.L., J.E.L., B.S.G., M.C., and D.C.D. N.J.S, A.D.E., J.R. Mascola, and G.G. supervised the study. B.W., B.J.D., J.R. Mascola, and G.G. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
Competing financial interests statement G.G., B.J.D., and A.D.E. declare competing financial interests in the form of a patent filed by the University of Texas at Austin.
Integrated supplementary information
Supplementary Figure 1 Map of pCT-VHVL-K1 native VH:VL display vector.
Natively-paired VH:VL sequences are cloned en masse into this vector for human antibody repertoire mining, and their corresponding Fabs are expressed on the yeast cell surface via galactose induction.
Supplementary Figure 2 Flow cytometry analysis of a panel of human anti-HA antibodies before and after display optimization, and of the 2 anti-EBOV antibodies and 1 anti-HIV-1 bNAb in the optimized system.
(a) Display of the six anti-HA antibodies listed in Figure 1c that did not functionally display in EBY100 (upper) and in AWY101 with LZ-forced Fab dimerization (lower). (b) Anti-EBOV antibodies c13c6 and KZ52 and anti-HIV-1 bNAb VRC34.01 displayed in the optimized system. For anti-HA antibodies, 100nM recombinant A/California/07/2009 HA was used to stain D1 H1-2, D1 H1-3/H3-3, D1 H1-53, D1 H1-12, and D1 H1-17/H3-14, and 100nM recombinant B/Brisbane/60/2008 HA was used to stain D1 Vic-8/Yama-20. 23 nM GPΔmuc-APC was used to stain c13c6 and KZ52; 50nM VRC34-epitope scaffold-FP-APC was used to stain VRC34.01. A representative profile from 5 (a) or 3 (b) independent experiments for each antibody is shown.
Supplementary Figure 3 Representative FACS gating strategy for EBOV library sorts.
Yeast cells were stained with 2 μg/ml anti-FLAG-FITC and 23 nM GPΔmuc-APC.
Supplementary Figure 4 Flow cytometry antigen binding profiles of monoclonal yeast populations expressing EBOV.YD.09-EBOV.YD.11, which were identified by single colony picking.
Yeast cells were stained with 2 μg/ml anti-FLAG-FITC and 23 nM GPΔmuc-APC.
Supplementary Figure 5 Biolayer interferometry response curves for human anti-EBOV antibodies from the plasmablast cognate VH:VL repertoire.
Binding was assessed against GPΔmuc. Global analyses were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
Supplementary Figure 6 Neutralization of EBOV GP pseudotype infection by human anti-EBOV antibodies.
% Infection is shown relative to the negative control antibody VRC01. Data are reported as average ± standard deviation for three technical replicates.
Supplementary Figure 7 Representative FACS gating strategy for HIV-1 library sorts.
Yeast cells were stained with 2 μg/ml anti-FLAG-FITC, 50 nM VRC34-epitope scaffold-FP-APC, and 50 nM VRC34-epitope scaffold-KO-PE for the isolation of HIV-1 fusion peptide-specific antibodies.
Supplementary Figure 8 Biolayer interferometry response curves for HIV-1 FP-specific antibodies from the B cell repertoire of an HIV-1 slow progressor.
The FP-scaffold protein was immobilized on the biosensor chip. Global analyses were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
Supplementary Figure 9 HIV-1 neutralization IC50 potency for natively paired VRC34 family antibodies discovered via yeast display.
Neutralization was determined against a panel of 22 viruses.
Supplementary Figure 10 Sequence alignments of HIV-1 FP-specific antibodies from the peripheral B cell repertoire of donor N123.
Mutations are colored in red.
Supplementary Figure 11 Representative FACS gating strategy for flu library screening.
Yeast cells were stained with 2 μg/ml anti-FLAG-FITC, and either 40nM A/Solomon Islands/3/2006 H1 HA (upper panel) or 40nM A/Wisconsin/67/2005 H3 HA (lower panel) for the isolation of H1 and H3-specific antibodies, respectively.
Supplementary Figure 12 Surface plasmon resonance binding curves for anti-HA antibodies from the B cell cognate VH:VL repertoire.
Representative binding curves from 3 independent experiments for each antibody are shown.
Supplementary Figure 13 Neutralization profiles of anti-HA antibodies isolated via yeast display.
CR9114 and CR6261 were included as positive and negative controls, respectively. Data are reported as average ± standard deviation from three technical replicates.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 2564 kb)
Supplementary Tables
Supplementary Tables 1–5 (PDF 425 kb)
Rights and permissions
About this article
Cite this article
Wang, B., DeKosky, B., Timm, M. et al. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat Biotechnol 36, 152–155 (2018). https://doi.org/10.1038/nbt.4052
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4052
This article is cited by
-
A fine-tuned yeast surface-display/secretion platform enables the rapid discovery of neutralizing antibodies against Clostridioides difficile toxins
Microbial Cell Factories (2023)
-
Insights into next generation sequencing guided antibody selection strategies
Scientific Reports (2023)
-
Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site
Nature Communications (2023)
-
Cell activation-based screening of natively paired human T cell receptor repertoires
Scientific Reports (2023)
-
Honing-in antigen-specific cells during antibody discovery: a user-friendly process to mine a deeper repertoire
Communications Biology (2022)