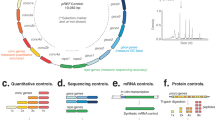
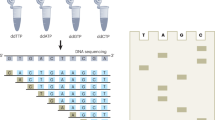
Abstract
The construction of synthetic biological systems involving millions of nucleotides is limited by the lack of high-quality synthetic DNA. Consequently, the field requires advances in the accuracy and scale of chemical DNA synthesis and in the processing of longer DNA assembled from short fragments. Here we describe a highly parallel and miniaturized method, called megacloning, for obtaining high-quality DNA by using next-generation sequencing (NGS) technology as a preparative tool. We demonstrate our method by processing both chemically synthesized and microarray-derived DNA oligonucleotides with a robotic system for imaging and picking beads directly off of a high-throughput pyrosequencing platform. The method can reduce error rates by a factor of 500 compared to the starting oligonucleotide pool generated by microarray. We use DNA obtained by megacloning to assemble synthetic genes. In principle, millions of DNA fragments can be sequenced, characterized and sorted in a single megacloner run, enabling constructive biology up to the megabase scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Endy, D. Foundations for engineering biology. Nature 438, 449–453 (2005).
Menzella, H.G. et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 23, 1171–1176 (2005).
Gibson, D.G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Carr, P.A. & Church, G.M. Genome engineering. Nat. Biotechnol. 27, 1151–1162 (2009).
Gao, X. et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 29, 4744–4750 (2001).
Singh-Gasson, S. et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17, 974–978 (1999).
Tian, J. et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054 (2004).
Porreca, G.J. et al. Multiplex amplification of large sets of human exons. Nat. Methods 4, 931–936 (2007).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Wicker, T. et al. 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7, 275 (2006).
Willenbrock, H. et al. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA 15, 2028–2034 (2009).
Stemmer, W.P., Crameri, A., Ha, K.D., Brennan, T.M. & Heyneker, H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 (1995).
Richmond, K.E. et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 32, 5011–5018 (2004).
Jefferson, R.A., Burgess, S.M. & Hirsh, D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83, 8447–8451 (1986).
Couteaudier, Y., Daboussi, M.J., Eparvier, A., Langin, T. & Orcival, J. The GUS gene fusion system (Escherichia coli beta-D-glucuronidase gene), a useful tool in studies of root colonization by Fusarium oxysporum. Appl. Environ. Microbiol. 59, 1767–1773 (1993).
Cline, J., Braman, J.C. & Hogrefe, H.H. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24, 3546–3551 (1996).
Carr, P.A. et al. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 32, e162 (2004).
Smith, J. & Modrich, P. Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl. Acad. Sci. USA 94, 6847–6850 (1997).
Bang, D. & Church, G.M. Gene synthesis by circular assembly amplification. Nat. Methods 5, 37–39 (2008).
Fuhrmann, M., Oertel, W., Berthold, P. & Hegemann, P. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage. Nucleic Acids Res. 33, e58 (2005).
Binkowski, B.F., Richmond, K.E., Kaysen, J., Sussman, M.R. & Belshaw, P.J. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res. 33, e55 (2005).
McKernan, K.J. et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 19, 1527–1541 (2009).
Kosuri, S. et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. advance online publication, doi:10.1038/nbt.1716 (28 November 2010).
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3, 545–550 (2006).
Acknowledgements
We thank B.A. Roe, F.Z. Najar and D.D. White for sequencing support, J. Jäger for technical consulting, and D. Summerer, T. Brefort, S. Kosuri and D. Levner for discussions and comments.
Author information
Authors and Affiliations
Contributions
M.M., P.F.S. and G.M.C. conceptualized the megacloning method and wrote the manuscript; M.M. designed and lead the study, wrote all algorithms for sequence design, data analysis, image conversion, image processing and microactuator control; M.M., N.K., N.S. acquired the used technology, set up the microactuator device and optical systems; N.S. designed the uidA genetic model; M.M., N.K., N.S., V.B. and P.H. designed and optimized molecular biological methods; C.F.S. and J.T.L. contributed to bead picking and engineering concepts; A.K. set up the statistical models and calculations; J.T.L. contributed to the design of molecular biological steps and the acquisition of sequencing samples; B.G. and F.B. evaluated and implemented necessary changes into the sample preparation and the sequencing process on the 454/Roche platform.
Corresponding author
Ethics declarations
Competing interests
M.M., N.K., N.S., V.B., J.T.L., P.F.S., A.K., C.F.S. and P.H. have potentially competing financial interests in companies affiliated to the febit group.
Supplementary information
Supplementary Text and Figures
Supplementary Data (PDF 3294 kb)
Rights and permissions
About this article
Cite this article
Matzas, M., Stähler, P., Kefer, N. et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat Biotechnol 28, 1291–1294 (2010). https://doi.org/10.1038/nbt.1710
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1710
This article is cited by
-
Purification of multiplex oligonucleotide libraries by synthesis and selection
Nature Biotechnology (2022)
-
An overview and future prospects of recombinant protein production in Bacillus subtilis
Applied Microbiology and Biotechnology (2021)
-
Whole genome engineering by synthesis
Science China Life Sciences (2018)
-
High-fidelity de novo synthesis of pathways using microchip-synthesized oligonucleotides and general molecular biology equipment
Scientific Reports (2017)
-
Whole genome synthesis: from poliovirus to synthetic yeast
Quantitative Biology (2017)