Abstract
Modern ray-finned fishes (Actinopterygii) comprise half of extant vertebrate species and are widely thought to have originated before or near the end of the Middle Devonian epoch (around 385 million years ago)1,2,3,4. Polypterids (bichirs and ropefish) represent the earliest-diverging lineage of living actinopterygians, with almost all Palaeozoic taxa interpreted as more closely related to other extant actinopterygians than to polypterids5,6,7,8,9,10. By contrast, the earliest material assigned to the polypterid lineage is mid-Cretaceous in age (around 100 million years old)11, implying a quarter-of-a-billion-year palaeontological gap. Here we show that scanilepiforms, a widely distributed radiation from the Triassic period (around 252–201 million years ago), are stem polypterids. Importantly, these fossils break the long polypterid branch and expose many supposedly primitive features of extant polypterids as reversals. This shifts numerous Palaeozoic ray-fins to the actinopterygian stem, reducing the minimum age for the crown lineage by roughly 45 million years. Recalibration of molecular clocks to exclude phylogenetically reassigned Palaeozoic taxa results in estimates that the actinopterygian crown lineage is about 20–40 million years younger than was indicated by previous molecular analyses1,2,3,4. These new dates are broadly consistent with our revised palaeontological timescale and coincident with an interval of conspicuous morphological and taxonomic diversification among ray-fins centred on the Devonian–Carboniferous boundary12,13,14. A shifting timescale, combined with ambiguity in the relationships of late Palaeozoic actinopterygians, highlights this part of the fossil record as a major frontier in understanding the evolutionary assembly of modern vertebrate diversity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
22 November 2017
Please see accompanying Erratum (http://doi.org/10.1038/nature25004). Nine Supplementary Data files were missing from the online version of this Letter; these have been added to the HTML.
References
Hurley, I. A . et al. A new time-scale for ray-finned fish evolution. Proc. R. Soc. Lond. B 274, 489–498 (2007)
Near, T. J. et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl Acad. Sci. USA 109, 13698–13703 (2012)
Broughton, R. E., Betancur- R, R., Li, C., Arratia, G. & Ortí, G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. http://dx.doi.org/10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e (2013)
Faircloth, B. C., Sorenson, L., Santini, F. & Alfaro, M. E. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS One 8, e65923 (2013)
Patterson, C. Morphology and interrelationships of primitive actinopterygian fishes. Am. Zool. 22, 241–295 (1982)
Gardiner, B. G. The relationships of the palaeoniscid fishes, a review based on new specimens of Mimia and Moythomasia from the Upper Devonian of Western Australia. Bull. Br. Mus. Nat. Hist. 37, 173–428 (1984)
Coates, M. I. Endocranial preservation of a Carboniferous actinopterygian from Lancashire, UK, and the interrelationships of primitive actinopterygians. Philos. Trans. R. Soc. Lond. B 354, 435–462 (1999)
Sallan, L. C. Major issues in the origins of ray-finned fish (Actinopterygii) biodiversity. Biol. Rev. Camb. Philos. Soc. 89, 950–971 (2014)
Xu, G.-H., Gao, K. Q. & Finarelli, J. A. A revision of the Middle Triassic scanilepiform fish Fukangichthys longidorsalis from Xinjiang, China, with comments on the phylogeny of the Actinopteri. J. Vertebr. Paleontol. 34, 747–759 (2014)
Friedman, M. The early evolution of ray-finned fishes. Palaeontology 58, 213–228 (2015)
Gayet, M., Meunier, F. J. & Werner, C. Diversification in Polypteriformes and special comparison with the Lepisosteiformes. Palaeontology 45, 361–376 (2002)
Sallan, L. C. & Coates, M. I. Styracopterid (Actinopterygii) ontogeny and the multiple origins of post-Hangenburg deep bodied fishes. Zool. J. Linn. Soc. 169, 156–199 (2013)
Sallan, L. C . & Friedman, M. Heads or tails: staged diversification in vertebrate evolutionary radiations. Proc. R. Soc. B http://dx.doi.org/10.1098/rspb.2011.2454 (2011)
Friedman, M. & Sallan, L. C. Five hundred million years of extinction and recovery: a Phanerozoic survey of large-scale diversity patterns in fishes. Palaeontology 55, 707–742 (2012)
Near, T. J. et al. Boom and bust: ancient and recent diversification in bichirs (Polypteridae: Actinopterygii), a relictual lineage of ray-finned fishes. Evolution 68, 1014–1026 (2014)
Goodrich, E. S. Polypterus, a palaeoniscid? Palaeobiologica 1, 87–91 (1928)
Allis, E. P. The cranial anatomy of Polypterus, with special reference to Polypterus bichir. J. Anat. 56, 189–294, 43 (1922)
Jessen, H. L. In Interrelationships of Fishes (eds Greenwood, P. H., Miles, R. S. & Patterson, C. ) 63–103 (Academic, 1973)
Grande, L. An empirical synthetic pattern study of gars (Lepisosteiformes) and closely related species, based mostly on skeletal anatomy. The resurrection of Holostei. Am. Soc. Ichthyol. Herpetol. Special Publication 6, 1–871 (2010)
Gardiner, B. G., Schaeffer, B. & Masserie, J. A. A review of the lower actinopterygian phylogeny. Zool. J. Linn. Soc. 144, 511–525 (2005)
Duthiel, D. B. The first articulated fossil cladistian: Serenoichthys kemkemensis, gen. et sp. nov., from the Cretaceous of Morocco. J. Vertebr. Paleontol. 19, 243–246 (1999)
Jollie, M. Development of the head and pectoral skeleton of Polypterus with a note on scales (Pisces: Actinopterygii). J. Zool. 204, 469–507 (1984)
Claeson, K. M., Bemis, W. E. & Hagadorn, J. W. New interpretations of the skull of a primitive bony fish Erpetoichthys calabaricus (Actinopterygii: Cladistia). J. Morphol. 268, 1021–1039 (2007)
Selezneva, A. A. Evenkia — Ancestor of Polypterus (Actinopterygii). Paleontol. J. 19, 1–6 (1985)
Sytchevskaya, E. K. in Mesozoic Fishes 2. Systematics and Fossil Record (eds Arratia, G. & Schultze H.-P. ) 445–468 (Dr. Friedrich Pfeil, 1999)
Xu, G.-H. & Gao, K.-Q. A new scanilepiform from the Lower Triassic of northern Gansu Province, China, and the phylogenetic relationships of non-teleostean Actinopterygii. Zool. J. Linn. Soc. 161, 595–612 (2011)
Giles, S ., Darras, L ., Clément, G ., Blieck, A . & Friedman, M. An exceptionally preserved Late Devonian actinopterygian provides a new model for primitive cranial anatomy in ray-finned fishes. Proc. R. Soc. Lond. B 282, 2015 1485 (2015)
Cloutier, R. & Arratia, G. In Recent Advances in the Origin and Early Radiation of Vertebrates (eds Arratia, G., Wilson, M. H. V. & Cloutier, R. ) 217–270 (Dr. Friedrich Pfeil, 2004)
Mickle, K. E., Lund, R. & Grogan, E. D. Three new palaeoniscoid fishes from the Bear Gulch Limestone (Serpukhovian, Mississippian) of Montana (USA) and the relationships of lower actinopterygians. Geodiversitas 31, 623–668 (2009)
Benton, M. J. et al. Constraints on the timescale of animal evolutionary history. Palaeo. Electronica 18.1.1FC (2015)
Britz, R. & Johnson, G. D. On the homology of the posteriormost gill arch in polypterids (Cladistia, Actinopterygii). Zool. J. Linn. Soc. 138, 495–503 (2011)
Swofford, D. L. PAUP*: Phylogenetic Analysis Using Parsimony (*And Other Methods) v.4.0b 10 (Sinauer Associates, 2003)
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001)
Wilkinson, M. Coping with missing entries in phylogenetic inference using parsimony. Syst. Biol. 44, 501–514 (1995)
Lloyd, G. T. Claddis: an R package for performing disparity and rate analysis on cladistic-type data sets. https://github.com/graemetlloyd/Claddis (GitHub, 2015)
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1.6. http://tree.bio.ed.ac.uk/software/tracer/ (2014)
Aberer, A. J., Krompass, D. & Stamatakis, A. Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst. Biol. 62, 162–166 (2013)
R Core Team. R: A language and environment for statistical computing. https://www.R-project.org/ (R Foundation for Statistical Computing, 2016)
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004)
Mark, A. Bell . geoscale: Geological Time Scale Plotting. R package version 2.0. https://CRAN.R-project.org/package=geoscale (2015)
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007)
Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006)
Giles, S. Xu, G.-H., Near, T. J. & Friedman, M. Fukangichthys: CT scan data and surface files from Middle Triassic fossil scanilepiform fish. https://doi.org/10.6084/m9.figshare.c.3814360 (2017)
Acknowledgements
Y.-M. Hou, D. Sykes and R. Summerfield assisted with CT scanning. C. Healy, Z. Johanson and M. M. Smith provided scans of Erpetoichthys. L. Sallan provided helpful discussion. N. Brocklehurst helped with R, and L. Parry assisted with R and MrBayes. S.G. was supported by a Junior Research Fellowship from Christ Church, Oxford, and a L’Oréal-UNESCO For Women in Science Fellowship. G.-H.X. was supported by the National Natural Science Foundation of China (41672001). T.J.N was supported by the National Science Foundation (ANT-134166) and the Bingham Oceanographic Fund from the Peabody Museum of Natural History, Yale University. M.F. was supported by a Philip Leverhulme Prize (PLP-2012-130) and Leverhulme Trust Project Grant (RPG-2012-65A).
Author information
Authors and Affiliations
Contributions
The project was conceived by M.F. CT scanning was carried out by S.G., G.-H.X. and M.F. S.G. segmented the CT data, conducted phylogenetic and leaf stability analyses and created the figures, with input from M.F. T.J.N. calculated divergence estimates. S.G. and M.F. wrote the manuscript, with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks W. Bemis and M. Coates for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
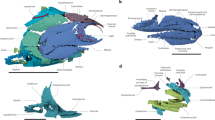
Extended Data Figure 1 Cranial anatomy of Fukangichthys longidorsalis based on high-resolution computed tomography.
a, Lateral view of whole skull (IVPP V4096.13). b, Lateral view of braincase, hyomandibula and lower jaw (IVPP V4096.6). c, Braincase in anterior view (IVPP V4096.6). d, Hyoid and branchial arches in dorsal view (IVPP V4096.13). e, Jaws and palate in ventral view (IVPP V4096.13). f, Left lateral view of whole skull (IVPP V4096.6). g, Right lateral view of whole skull (IVPP V4096.6). bb, basibranchial; clav, clavicle; clth, cleithrum; dsph, dermosphenotic; eb, epibranchial; hh, hypohyal; jug, jugal; la, lachrymal; l.ex, lateral extrascapular; m.ex, median extrascapular; opm, operculum; pb, pharyngobranchial; pq, palatoquadrate; prop, preoperculum; pt, posttemporal; qj, quadratojugal; sop, suboperculum; spcl, supracleithrum; sr, skull roof. Mouldic portion of lower jaw shaded. Other abbreviations and colours as in Fig. 1. Scale bars, 5 mm (a, b, d–g); 2 mm (c).
Extended Data Figure 2 Photographs of Fukangichthys longidorsalis specimens examined in this study.
a, IVPP V4096.13 in left lateral view. b, IVPP V4096.13 in ventral view. c, IVPP V4096.13 in dorsal view. d, IVPP V4096.6 in left lateral view. e, IVPP V4096.6 in right lateral view. f, IVPP V4096.6 in dorsal view. Scale bar, 10 mm.
Extended Data Figure 3 Interpretive drawings of comparative cranial anatomy of Fukangichthys longidorsalis (IVPP V4096.6 and IVPP V4096.13) and Erpetoichthys calabaricus (BMNH 2016.9.22.3) based on high-resolution computed tomography.
a–e, Fukangichthys longidorsalis. f–j, Erpetoichthys calabaricus. a, Lateral view of whole skull (IVPP V4096.13). b, Lateral view of braincase, hyomandibula and lower jaw (IVPP V4096.6). c, Braincase in anterior view (IVPP V4096.6). d, Hyoid and branchial arches in dorsal view (IVPP V4096.13). e, Jaws and palate in ventral view (IVPP V4096.13). f, Left lateral view of whole skull (IVPP V4096.6). g, Right lateral view of whole skull (IVPP V4096.6). Scale bars, 5 mm (a, b, e–g, j); 2 mm (c, d, h, i).
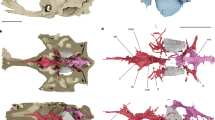
Extended Data Figure 4 Comparative palatal anatomy of Fukangichthys longidorsalis (IVPP V4096.6 and IVPP V4096.13) and Erpetoichthys calabaricus (BMNH 2016.9.22.3) based on high-resolution computed tomography.
a–c, e, Fukangichthys longidorsalis. d, f, Erpetoichthys calabaricus. a, Medial view of left palate (IVPP V4096.13). b, Medial view of right palate (IVPP V4096.13). c, Medial view of left palate (IVPP V4096.6). d, Medial view of left palate. e, Anterolateral view of left palate (IVPP V4096.13). e, Anterolateral view of left palate. qu, quadrate. Scale bars, 5 mm.
Extended Data Figure 5 Comparative hyoid and branchial anatomy of Fukangichthys longidorsalis (IVPP V4096.6 and IVPP V4096.13) and Erpetoichthys calabaricus (BMNH 2016.9.22.3) based on high-resolution computed tomography.
a, d, g, j, Erpetoichthys calabaricus. b, c, e, f, h, i, k–m, Fukangichthys longidorsalis. a, Braincase, palate, mandibular arch, hyoid arch and ventral portion of branchial arch in ventral view. b, Braincase, palate, mandibular arch, hyoid arch and ventral portion of branchial arch in ventral view (IVPP V4096.6). c, Braincase, palate, mandibular arch, hyoid arch and branchial arch in ventral view (IVPP V4096.13). d, Ventral portion of hyoid and branchial arches in ventral view. e, Ventral portion of hyoid and branchial arches in ventral view (IVPP V4096.6). f, Branchial arches and ventral portion of hyoid arch in ventral view (IVPP V4096.13). g, Ventral portion of hyoid and branchial arches in dorsal view. h, Ventral portion of hyoid and branchial arches in dorsal view (IVPP V4096.6). i, Branchial arches and ventral portion of hyoid arch in ventral view (IVPP V4096.13). j, Hyoid arch and ventral portion of branchial arches in lateral view. k, Hyoid arch and ventral portion of branchial arches in lateral view (IVPP V4096.6). l, Hyoid and branchial arches in lateral view (IVPP V4096.13). m, Close-up of uncinated process of epibranchial. ahy, groove for afferent hyoid artery; ih, interhyal; up, uncinate process. Colours as in Fig. 1. Scale bars 5 mm (a–l); 1 mm (m).
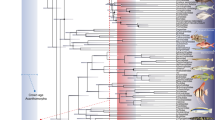
Extended Data Figure 6 Results of phylogenetic analyses.
a, Strict consensus of the 14,450 shortest trees (1,347 steps) for 93 taxa and 265 equally weighted characters. Digits above nodes indicate Bremer decay indices above 1. Digits below nodes indicate percentage bootstrap support above 50%. b, Adams consensus tree of the 14,450 shortest trees (1,347 steps) for 93 taxa and 265 equally weighted characters. Scanilepids and polypterids shown in bold.
Extended Data Figure 7 Results of phylogenetic analyses.
Agreement subtree of the 14,450 shortest trees (1,347 steps) for 93 taxa and 265 equally weighted characters. Scanilepids and polypterids shown in bold. 78 of 93 taxa are included, and the following taxa are pruned from the tree: Beagiascus pulcherrimus, Beishanichthys brevicaudalis, Birgeria groenlandica, Cosmoptychius striatus, Cyranorhis bergeraci, Guiyu oneiros, Howqualepis rostridens, Lawrenciella schaefferi, Mimipiscis toombsi, Onychodus jandemarrai, Platysomus superbus, Psarolepis romeri, Scanilepis dubia, Tanaocrossus kalliokoskii and Wendyichthys dicksoni.
Extended Data Figure 8 Results of Bayesian analyses.
a, Combined morphological and molecular dataset. Terminals in blue are coded for both molecular and morphological data. Scanilepids and polypterids shown in bold. b, Morphological-only dataset. Numbers at nodes represent posterior probability support; asterisks represent a posterior probability of 1.
Extended Data Figure 9 Endocast and bony labyrinth of Erpetoichthys calabaricus (BMNH 2016.9.22.3) showing vestigial lateral cranial canal.
a, Dorsal view. b, Lateral view. c, Transverse tomograph through otic region. a.amp, ampulla of the anterior semicircular canal; asc, posterior semicircular canal; cr.cav, cranial cavity; h.amp, ampulla of the horizontal semicircular canal; hsc, horizontal semicircular canal; lcc, lateral cranial canal; psc, posterior semicircular canal; sac, sacculus. Scale bars, 5 mm.
Extended Data Figure 10 Leaf stability analyses.
a, Raw leaf stability plotted against taxon age (same data as in Fig. 3a). b, Raw leaf stability plotted against taxon age. Error bars represent s.d. (same data as in Fig. 3a). c, Residuals from a linear regression of stability against taxon incompleteness, plotted against taxon age (same data as in Fig. 3b). Taxa identified in Supplementary Table 2.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Tables 1-2, Phylogenetic analysis including anatomical character list and additional references. (PDF 2114 kb)
Supplementary Data
This file contains a large format PDF of a single most parsimonious tree showing all character optimisations. (PDF 477 kb)
Supplementary Data
This file contains Supplementary Data files 1-9 and a Supplementary Data guide – this file was added 22nd November 2017, see Erratum 10.1038/nature25004 for details. (ZIP 722 kb)
Rights and permissions
About this article
Cite this article
Giles, S., Xu, GH., Near, T. et al. Early members of ‘living fossil’ lineage imply later origin of modern ray-finned fishes. Nature 549, 265–268 (2017). https://doi.org/10.1038/nature23654
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23654
This article is cited by
-
Phylogenetic congruence, conflict and consilience between molecular and morphological data
BMC Ecology and Evolution (2023)
-
Independent rediploidization masks shared whole genome duplication in the sturgeon-paddlefish ancestor
Nature Communications (2023)
-
Exceptional fossil preservation and evolution of the ray-finned fish brain
Nature (2023)
-
Fish fossil unfolds clues to vertebrate brain evolution
Nature (2023)
-
Ancient fish lineages illuminate toll-like receptor diversification in early vertebrate evolution
Immunogenetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.