Abstract
Similar to resting mature B cells, where the B-cell antigen receptor (BCR) controls cellular survival1,2,3, surface BCR expression is conserved in most mature B-cell lymphomas. The identification of activating BCR mutations and the growth disadvantage upon BCR knockdown of cells of certain lymphoma entities has led to the view that BCR signalling is required for tumour cell survival4,5,6,7. Consequently, the BCR signalling machinery has become an established target in the therapy of B-cell malignancies8,9. Here we study the effects of BCR ablation on MYC-driven mouse B-cell lymphomas and compare them with observations in human Burkitt lymphoma. Whereas BCR ablation does not, per se, significantly affect lymphoma growth, BCR-negative (BCR−) tumour cells rapidly disappear in the presence of their BCR-expressing (BCR+) counterparts in vitro and in vivo. This requires neither cellular contact nor factors released by BCR+ tumour cells. Instead, BCR loss induces the rewiring of central carbon metabolism, increasing the sensitivity of receptor-less lymphoma cells to nutrient restriction. The BCR attenuates glycogen synthase kinase 3 beta (GSK3β) activity to support MYC-controlled gene expression. BCR− tumour cells exhibit increased GSK3β activity and are rescued from their competitive growth disadvantage by GSK3β inhibition. BCR− lymphoma variants that restore competitive fitness normalize GSK3β activity after constitutive activation of the MAPK pathway, commonly through Ras mutations. Similarly, in Burkitt lymphoma, activating RAS mutations may propagate immunoglobulin-crippled tumour cells, which usually represent a minority of the tumour bulk. Thus, while BCR expression enhances lymphoma cell fitness, BCR-targeted therapies may profit from combinations with drugs targeting BCR− tumour cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lam, K. P., Kühn, R. & Rajewsky, K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 90, 1073–1083 (1997)
Kraus, M., Alimzhanov, M. B., Rajewsky, N. & Rajewsky, K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell 117, 787–800 (2004)
Srinivasan, L. et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139, 573–586 (2009)
Davis, R. E. et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 (2010)
Schmitz, R. et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012)
Young, R. M., Shaffer, A. L. III, Phelan, J. D. & Staudt, L. M. B-cell receptor signaling in diffuse large B-cell lymphoma. Semin. Hematol. 52, 77–85 (2015)
Refaeli, Y. et al. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol. 6, e152 (2008)
Young, R. M. & Staudt, L. M. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat. Rev. Drug Discov. 12, 229–243 (2013)
Wilson, W. H. et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat. Med. 21, 922–926 (2015)
Kovalchuk, A. L. et al. Burkitt lymphoma in the mouse. J. Exp. Med. 192, 1183–1190 (2000)
Sander, S. et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22, 167–179 (2012)
Schmidt-Supprian, M. & Rajewsky, K. Vagaries of conditional gene targeting. Nat. Immunol. 8, 665–668 (2007)
Levit-Zerdoun, E. et al. Survival of Igα-deficient mature B cells requires BAFF-R function. J. Immunol. 196, 2348–2360 (2016)
Pietzke, M. & Kempa, S. Pulsed stable isotope-resolved metabolomic studies of cancer cells. Methods Enzymol. 543, 179–198 (2014)
Gold, M. R. et al. The B cell antigen receptor activates the Akt (protein kinase B)/glycogen synthase kinase-3 signaling pathway via phosphatidylinositol 3-kinase. J. Immunol. 163, 1894–1905 (1999)
Cohen, P. & Frame, S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 (2001)
Ring, D. B. et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52, 588–595 (2003)
Otipoby, K. L. et al. The B-cell antigen receptor integrates adaptive and innate immune signals. Proc. Natl Acad. Sci. USA 112, 12145–12150 (2015)
Corso, J. et al. Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc. Natl Acad. Sci. USA 113, 5688–5693 (2016)
Sabò, A. et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014)
Smith, G. et al. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours – implications for personalised cancer medicine. Br. J. Cancer 102, 693–703 (2010)
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012)
Rowland, S. L., DePersis, C. L., Torres, R. M. & Pelanda, R. Ras activation of Erk restores impaired tonic BCR signaling and rescues immature B cell differentiation. J. Exp. Med. 207, 607–621 (2010)
Teodorovic, L. S. et al. Activation of Ras overcomes B-cell tolerance to promote differentiation of autoreactive B cells and production of autoantibodies. Proc. Natl Acad. Sci. USA 111, E2797–E2806 (2014)
Wang, T. et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903 (2017)
Schmitz, R., Ceribelli, M., Pittaluga, S., Wright, G. & Staudt, L. M. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb. Perspect. Med. 4, a014282 (2014)
Casola, S. et al. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5, 317–327 (2004)
Peitz, M., Pfannkuche, K., Rajewsky, K. & Edenhofer, F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl Acad. Sci. USA 99, 4489–4494 (2002)
Di Micco, R. et al. DNA damage response activation in mouse embryonic fibroblasts undergoing replicative senescence and following spontaneous immortalization. Cell Cycle 7, 3601–3606 (2008)
Gao, Y. et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404, 897–900 (2000)
Cheung, W. C. et al. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J. Clin. Invest. 113, 1763–1773 (2004)
Caganova, M. et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 123, 5009–5022 (2013)
Kempa, S. et al. An automated GCxGC-TOF-MS protocol for batch-wise extraction and alignment of mass isotopomer matrixes from differential 13C-labelling experiments: a case study for photoautotrophic-mixotrophic grown Chlamydomonas reinhardtii cells. J. Basic Microbiol. 49, 82–91 (2009)
Pietzke, M., Zasada, C., Mudrich, S. & Kempa, S. Decoding the dynamics of cellular metabolism and the action of 3-bromopyruvate and 2-deoxyglucose using pulsed stable isotope-resolved metabolomics. Cancer Metab. 2, 9 (2014)
Kopka, J. et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 (2005)
Kuich, P. H., Hoffmann, N. & Kempa, S. Maui-VIA: a user-friendly software for visual identification, alignment, correction, and quantification of gas chromatography-mass spectrometry data. Front. Bioeng. Biotechnol. 2, 84 (2015)
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013)
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97 (2016)
Trifonov, V., Pasqualucci, L., Tiacci, E., Falini, B. & Rabadan, R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst. Biol. 7 (Suppl. 2), S2 (2013)
Vermi, W. et al. Cutaneous distribution of plasmacytoid dendritic cells in lupus erythematosus. Selective tropism at the site of epithelial apoptotic damage. Immunobiology 214, 877–886 (2009)
Richter, J. et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 44, 1316–1320 (2012)
van Dongen, J. J. et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17, 2257–2317 (2003)
Acknowledgements
We thank members of the Casola laboratory, A. Ciliberto, K. L. Otipoby and R. Küppers for discussions; A. Ciliberto and N. Bolli for reading the manuscript; D. Wang, M. Caganova, L. Duarte and S. Ronzoni for preliminary work and technical assistance; F. D’Adda di Fagagna for reagents. We acknowledge the German International Cancer Genome Consortium (ICGC) Molecular Mechanisms in Malignant Lymphoma by Sequencing (MMML-Seq) and Molecular Mechanisms in Malignant Lymphoma (MMML) networks. This work was supported by the Italian Association for Cancer Research, the Giovanni Armenise/Harvard Foundation (to S.C.), the National Institutes of Health (to K.R.) and the European Research Council (to S.C. through FIRB IDEAS, and to K.R., Advanced Grant 268921). R.S. was supported by ICGC MMML-Seq grant 01KU1002, ICGC-DE-Mining grant 01KU1505G and e:BIO MMML-MYC-SYS grant 036166, as well as by the KinderKrebsInitiative Buchholz/Holm-Seppensen. G.V. and V.P were supported by the U. Veronesi Foundation. S.L. and M.B. were supported by Fondazione Beretta.
Author information
Authors and Affiliations
Contributions
M.S. and F.Z. contributed equally to this work. G.V. performed most experiments. S.R. made initial observations. M.S. performed CAL-101 lymphoma sensitivity assays, in vivo transplantations and biochemical analyses. M.S. analysed Xbp1 expression with E.V.A. F.Z. performed BCR complementation studies and bioinformatics analyses on transcriptome and exome sequencing data. C.Z. and M.S. performed metabolic labelling of lymphomas. C.Z. and S.K. analysed and interpreted metabolomic data. L.Pe. investigated the role of cell contact and soluble factors in the counter-selection of BCR− tumour cells. V.P. performed Burkitt lymphoma immunoglobulin gene rearrangement analyses and Ras complementation assays. S.L., M.B., M.P. and F.F. performed immunohistochemical analyses and interpreted the results. A.H., U.P. and R.S. analysed RAS genes in Burkitt lymphoma. A.K.L., L.Pa. and R.R. assisted with exome sequencing analyses. S.C. and K.R. conceived the project. S.C. designed the experiments and interpreted the results with G.V. S.C. wrote and edited the manuscript with help from K.R. and G.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. Burger, A. Schaffer and A. Thomas-Tikhonenko for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
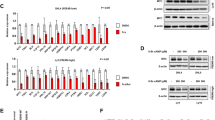
Extended Data Figure 1 Characterization of λ-MYC;B1-8f primary B-lymphoma cells.
a, The conditional λ-MYC;B1-8f mouse B-lymphoma model. b, Immunophenotypic characterization of B220+-gated tumour B cells from a representative λ-MYC;B1-8f tumour-bearing animal (thick black line). Comparison with splenic B220+-gated B cells from wild-type mice is shown (filled grey line). Experiments are representative of three lymphomas.
Extended Data Figure 2 Acute BCR ablation leaves the growth of λ-MYC lymphoma cells unperturbed.
a, In vitro growth curves of BCR+ and BCR− lymphoma clones grown in isolation, established from TAT-Cre-transduced λ-MYC;B1-8f primary lymphomas (n = 2). b, In vitro growth curves of representative pools of BCR+ and BCR− cells purified from TAT-Cre-transduced lymphomas (n = 2), grown in isolation. c, Kaplan–Meier curves of immunocompetent mice (BCR+, n = 7; BCR−, n = 8) transplanted with BCR+ or BCR− tumour clones established from a primary λ-MYC;B1-8f lymphoma (log-rank (Mantel–Cox) test: ***P < 0.001). d, IgM status in BCR+ (top) and BCR− (bottom) tumour clones from representative lymphomas, analysed before transplantation (Pre), or retrieved from the spleen of tumour-bearing recipients, 25 days after the transfer (Post). Frequencies of gated B220+ FSChi tumour B cells are indicated within plots. Experiments were performed with cells of three (a–d) lymphomas and data are representative of at least two experiments per tumour. Data represent mean values ± s.e.m. of three or more technical replicates (a, b).
Extended Data Figure 3 Effects of BCR ablation on competitive growth of MYC-transformed B cells.
a, IgH Southern blot analysis of representative λ-MYC;B1-8f lymphoma cells analysed at the indicated days of in vitro culture after TAT-Cre transduction. Bands correspond, respectively, to conditional (B1-8f) and Cre-recombined (B1-8∆) B1-8 alleles, and to the IgH germline (GL) allele. b, Average frequency of BCR+ and BCR− lymphoma cells assessed by flow cytometry at the onset, and 4 days after in vitro competition of representative lymphoma clones (n = 4). c, In vitro competitions of representative pools of BCR+ and BCR− lymphoma cells from three lymphomas followed for 6 days. Frequencies of BCR+ and BCR− lymphoma cells are represented. d, IgH Southern blot analysis of purified BCR+ and BCR− lymphoma pools, before (Day 0) transplantation into recipients as a 1:1 mixture, and in the spleen of tumour-bearing animals 14 days later. Tail-tip DNA from B1-8f homozygous mice (B1-8f/f) was included in the analysis as control. e, Representative flow cytometric analysis of CFSE-labelled BCR+/BCR− tumour mixtures from the indicated lymphomas before transplantation (Pre-injection), and present in the bone marrow (BM) of recipient animals 2 h after injection. Frequencies of gated FSChi CD19+ CFSE+ transplanted tumour cells (n = 2 per tumour) are shown. f, Strategy of IgH complementation in BCR− lymphoma cells. g, Average frequency of retrovirally complemented (GFP+) tumour B cells with the indicated surface BCR phenotype, before injection (Day 0) of IgMbGFP+/IgM−GFP+ tumour cell mixtures (2487, n = 2), and retrieved from the bone marrow and spleen (SPL) of transplanted mice 27 days after transfer. h, Quantification by qRT–PCR of spliced Xbp1 (Xbp1s) transcripts in pools of BCR+ and BCR− lymphoma cells purified from acutely transduced lymphomas grown in isolation. Data are relative to total Xbp1 (Xbp1t) messenger RNA levels. Representative BCR+ lymphoma cells treated with DTT to trigger the unfolded protein response were used as positive control. Experiments were performed on one (a, d, f, g), two (b, e) or more (c, h) lymphomas, and reproduced once (a, d), twice (e–h) or more (b, c). Boxes represent the 25th–75th percentiles and median (horizontal line), whiskers correspond to the 1.5 interquartile range added to the 75th percentile, or subtracted to the 25th percentile, respectively (b, c). Unpaired t-test (b, c) ***P < 0.001.
Extended Data Figure 4 BCR loss impairs cell-cycle progression and survival of MYC-transformed B cells during competition with BCR+ tumour cells.
a, Summary of cell-cycle distribution analysis data (n = 12) on BCR+ and BCR− lymphoma cells from four tumours grown in isolation. b, Representative flow cytometric determination of caspase activation (VAD-FMK+) among gated IgM+ and IgM− B-lymphoma cells (2676), retrieved after 5 days of in vitro competition. Numbers indicate frequency of VAD-FMK+ cells. c, Frequencies of VAD-FMK+ apoptotic lymphoma cells in BCR+ and BCR− populations of the indicated lymphomas, retrieved from days 4/5 of in vitro competition. d, Average frequency of apoptotic cells with subG1 DNA content among BCR− tumour B cells from four lymphomas (2646, 2676, 2564 and 2487), grown, respectively, in competition (n = 20) and isolation (n = 11). e, Numbers of BCR+ and BCR− tumour B cells measured from matched Transwell compartments were used to determine BCR−/BCR+ ratios at the indicated days of in vitro competitions. Clones from two lymphomas were analysed. f, Growth curves of BCR− lymphoma cells from two tumours (2646 and 2567) cultured in isolation for the indicated days in the presence of 50% conditioned medium/supernatant (SN) harvested from cultures of BCR+ and BCR− cells grown in isolation or from 1:1 mixtures (SN BCR+/−). Mean values ± s.e.m. of three technical replicates are shown. Experiments were performed on two (e) or more (a–d, f) lymphomas, and reproduced once (b, c, f), twice (e) or more (a, d) for each tumour. Boxes represent the 25th–75th percentiles and median (horizontal line), whiskers correspond to the 1.5 interquartile range added to the 75th percentile, or subtracted to the 25th percentile, respectively (a, d). Unpaired t-test (a, c, d), ***P < 0.001, NS, not significant.
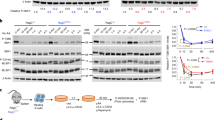
Extended Data Figure 5 BCR controls lymphoma cell fitness through GSK3β inactivation.
a, Representative flow cytometric analysis of BCR+/BCR− tumour mixtures (2676) cultured in vitro for 7 days in the presence of vehicle (DMSO) or CHIR99021. Numbers indicate frequency of cells within the corresponding quadrants. b, Frequencies of BCR− cells obtained from BCR+/BCR− tumour mixtures from two lymphomas, grown in vitro in the absence (−) or presence (+) of CHIR99021. Each symbol represents the average value of three measurements (unpaired t-test, **P < 0.01). c, Quantification by immunoblotting analysis of GSK3β levels in lymphoma cells (2567) after lentiviral expression of control (CTRL) or GSK3β targeting shRNAs (sh1 and sh2). Numbers indicate normalized GSK3β levels relative to control infected cells. d, Flow cytometric quantification of GSK3β levels in lymphoma cells (2567) after GSK3β knockdown using the indicated shRNAs. e, Histogram of the variation in BCR− tumour cell frequency 3 days after in vitro competition of BCR+/BCR− tumour cell mixtures (2567) infected with viruses expressing the indicated GSK3β-specific shRNAs, or with control virus. f, Effects of concomitant pharmacological inhibition of PI3Kδ and GSK3β on in vitro growth of BCR+ lymphoma cells (2646). Data refer to numbers (± s.e.m.) of treated tumour B cells relative to control cells grown in medium-containing carrier (DMSO). g, Frequency of viable (propidium-iodide-negative, PI−) lymphoma cells (± s.e.m.) from a representative tumour (2646) grown in vitro under concomitant inhibition of GSK3β (CHIR99021) and/or PI3Kδ (CAL-101). Frequencies are relative to vehicle (DMSO)-treated tumour cells. Unpaired t-test, ***P < 0.001). Data represent, respectively, one (c–g) and two (a, b) lymphomas, and were reproduced once (c, d), twice (e) or more (a, b, f, g) for each tumour.
Extended Data Figure 6 Gene networks regulated by the BCR–GSK3β axis in MYC-driven B lymphomas.
a, Number of genes differentially expressed (fold change > 1.3) between BCR+ and BCR− lymphoma cells. b, Number of genes whose expression is corrected (>30%) in BCR− tumour cells of three lymphomas retrieved from in vitro competitions treated with CHIR99021. c, Hierarchical clustering and heat map of genes controlled by the BCR through GSK3β inhibition in three lymphomas. Expression values are relative to the median expression value across all samples (log2 scale). d, Venn diagram of the number of genes differentially expressed between BCR+ and BCR− tumour B cells of three lymphomas (fold change > 1.3), which were corrected by pharmacological GSK3β inhibition. e, Top 15 ENCODE Transcription Factor ChIP-seq gene-set libraries significantly enriched (Fisher’s exact test, adjusted P < 0.05) for genes controlled by the BCR (fold change > 1.3) through GSK3β inhibition in three lymphomas (2646, n = 1,290; 2564, n = 750; 2676, n = 375), as revealed by the Enrichr software tool. Bioinformatic analyses were performed on transcriptome data obtained from tumour cells of three lymphomas purified from ongoing in vitro competitions (a–e).
Extended Data Figure 7 MYC-driven B-lymphoma cells select Ras mutations to bypass BCR requirement for optimal fitness.
a, Flow cytometric analysis of a representative BCR− clonal variant (2646 clone 7) resisting in vitro competition with BCR+ tumour cells (top), analysed at the indicated days after onset of competition. As a comparison, BCR− lymphoma cells succumbing to competition with their BCR+ counterparts were analysed (bottom). Numbers indicate frequency of viable tumour cells. b, Representative cell-cycle distribution analysis of, respectively, BCR+, BCR− and BCR-independent lymphoma clones (2646). Numbers indicate frequency of boxed cells. c, Cell-cycle distribution analysis of BCR+ and BCR-independent lymphoma clones from two tumours after 4 days of in vitro competition. d, Comparison of in vitro growth curves between BCR+ and BCR-independent lymphoma clones from a representative tumour (2646) grown in isolation. Mean values ± s.e.m of three replicates are shown. e, Numbers of BCR+ and BCR-independent lymphoma cells (2646) grown in isolation for 4 days in the presence of CAL-101. Data are relative to vehicle (DMSO)-treated control tumour cells. f, Frequency of viable (propidium-iodide-negative, PI−) BCR+ and BCR-independent tumour cells from representative lymphoma clones (2646) treated in vitro for 4 days with the indicated doses of CAL-101. Percentages of viable tumour cells are relative to vehicle-treated controls (DMSO). g, Venn diagram of number of differentially expressed genes between the indicated tumour subset comparisons of a representative lymphoma (2676). h, Gene categories significantly enriched (Fisher’s exact test, q < 0.05) among, respectively, upregulated (grey bars) and downregulated (black bars) genes in BCR-independent lymphoma cells compared with receptor-less tumour cells succumbing to the BCR+ counterparts (2646 and 2676). Dashed lines indicate threshold of significance (adjusted P = 0.05). i, Heat map of quantities of selected 13C-labelled metabolites measured, respectively, in BCR+, BCR− and BCR-independent lymphomas through pSIRM, derived from the catabolism of exogenous glutamine (Gln). j, Overlap of MYC target genes regulated by the BCR (BCR− versus BCR+) with those distinguishing BCR− lymphoma cells from BCR-independent tumour variants. k, Heat map of transcript levels of MYC target genes controlled by the BCR in lymphoma cells, measured in BCR-independent lymphoma cells. Expression values are relative to the median expression value across all samples (log2 scale). l, Sanger sequencing validation of Nras and Kras mutant alleles in BCR-independent clones established from three primary lymphomas. Arrows indicate single-nucleotide variants. m, Immunoblotting analysis of phospho-ERK (T202/Y204) levels in BCR+, BCR− and BCR-independent clones isolated from three lymphomas, including one lacking Ras mutations (2676 clone 1). n, Effect of activated RAS complementation on the in vitro out-competition of BCR− tumour B cells from three lymphomas analysed at the indicated days after TAT-Cre transduction. Competitions with BCR+/BCR− tumour mixtures infected with an empty virus (EV) acted as control. o, Growth curves of representative BCR-independent lymphoma cells cultured for 4 days in presence of the MEK inhibitor U0126 (10 μM), or vehicle (DMSO) (two-way analysis of variance, *P < 0.05; **P < 0.01). Mean values ± s.e.m of three replicates are shown. Experiments were performed on one (e, f), two (b–c, g–k, o) or three (a, d, l, m, n) independent lymphomas, and reproduced twice (b–f, m–o) or more (a) for each tumour.
Extended Data Figure 8 A subset of Burkitt lymphoma cases consists of Ig-negative lymphoma cells.
a, Immunohistochemical analysis of IgM (brown) and c-MYC (blue) expression in tissue sections of representative Burkitt lymphoma cases classified, respectively, as type 1, type 2 and type 3 tumours according to the frequency of IgM− tumour cells. Scale bar, 50 μm. b, Immunohistochemical validation of BCR negativity in representative cases of type 1, type 2 and type 3 Burkitt lymphoma. Tumour sections were immunostained for IgA, IgG and IgD isotypes. Staining for TdT expression was performed to exclude a progenitor B-cell origin of the tumours. Sections of human intestinal lamina propria were included as positive controls for IgA and sections of spleen as positive controls for IgG and IgD, respectively. A section from infiltrated testis with lymphoblastic B-cell lymphoma was used as positive control for TdT staining. Scale bar, 50 μm. c, Nucleotide sequence of clonal IGLV (lambda) and IGHV gene rearrangements amplified by PCR from a Burkitt lymphoma case consisting primarily of BCR− cells (case 4), as revealed by Sanger sequencing. Tumour IG V gene sequences are matched to the closest germline V gene identified by IgBLAST. Dots represent shared nucleotides. Asterisk (*) refers to a termination codon. Amino-acid changes resulting from nucleotide substitutions are shown in magenta. d, Sanger sequencing of a heterozygous NRASG12D hotspot mutation (arrow) identified in tumour material from a Burkitt lymphoma case with IgM-negative tumour B cells.
Supplementary information
Supplementary Figure
This file contains the uncropped blots with size marker indications. (PDF 940 kb)
Supplementary Data
This file contains Supplementary Tables 1-12. (XLSX 2788 kb)
Rights and permissions
About this article
Cite this article
Varano, G., Raffel, S., Sormani, M. et al. The B-cell receptor controls fitness of MYC-driven lymphoma cells via GSK3β inhibition. Nature 546, 302–306 (2017). https://doi.org/10.1038/nature22353
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22353
This article is cited by
-
Chronic lymphocytic leukemia patient-derived xenografts recapitulate clonal evolution to Richter transformation
Leukemia (2024)
-
Emerging entities: high-grade/large B-cell lymphoma with 11q aberration, large B-cell lymphoma with IRF4 rearrangement, and new molecular subgroups in large B-cell lymphomas. A report of the 2022 EA4HP/SH lymphoma workshop
Virchows Archiv (2023)
-
Intracellular osteopontin protects from autoimmunity-driven lymphoma development inhibiting TLR9-MYD88-STAT3 signaling
Molecular Cancer (2022)
-
Fatty acid metabolism in aggressive B-cell lymphoma is inhibited by tetraspanin CD37
Nature Communications (2022)
-
Detection of early seeding of Richter transformation in chronic lymphocytic leukemia
Nature Medicine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.