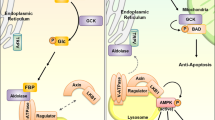
Abstract
The ability to sense and respond to fluctuations in environmental nutrient levels is a requisite for life. Nutrient scarcity is a selective pressure that has shaped the evolution of most cellular processes. Different pathways that detect intracellular and extracellular levels of sugars, amino acids, lipids and surrogate metabolites are integrated and coordinated at the organismal level through hormonal signals. During food abundance, nutrient-sensing pathways engage anabolism and storage, whereas scarcity triggers homeostatic mechanisms, such as the mobilization of internal stores through autophagy. Nutrient-sensing pathways are commonly deregulated in human metabolic diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wu, G. & Morris, S. M. Arginine metabolism: nitric oxide and beyond. Biochem. J. 336, 1–17 (1998).
Reeds, P. J. Dispensable and indispensable amino acids for humans. J. Nutr. 130, 1835S–1840S (2000).
Richieri, G. V. & Kleinfeld, A. M. Unbound free fatty acid levels in human serum. J. Lipid Res. 36, 229–240 (1995).
Itoh, Y. et al. Free fatty acids regulate insulin secretion from pancreatic β cells through GPR40. Nature 422, 173–176 (2003).
Hirasawa, A. et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nature Med. 11, 90–94 (2005).
Oh, D. Y. et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 (2010).
Ichimura, A. et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 483, 350–354 (2012).
Oh, D. Y. et al. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nature Med. 20, 942–947 (2014).
Pepino, M. Y., Kuda, O., Samovski, D. & Abumrad, N. A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 34, 281–303 (2014).
Laugerette, F. et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 115, 3177–3184 (2005).
Cartoni, C. et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. J. Neurosci. 30, 8376–8382 (2010).
Martin, C. et al. The lipid-sensor candidates CD36 and GPR120 are differentially regulated by dietary lipids in mouse taste buds: impact on spontaneous fat preference. PLoS ONE 6, e24014 (2011).
Pepino, M. Y., Love-Gregory, L., Klein, S. & Abumrad, N. A. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 53, 561–566 (2012).
Brown, M. S. & Goldstein, J. L. A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 (1986).
Brown, A. J., Sun, L., Feramisco, J. D., Brown, M. S. & Goldstein, J. L. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10, 237–245 (2002). This paper demonstrates the functional regulation of SCAP-protein conformation by cholesterol levels within the ER membrane, providing strong support for its cholesterol-sensing ability.
Radhakrishnan, A., Sun, L.-P., Kwon, H. J., Brown, M. S. & Goldstein, J. L. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell 15, 259–268 (2004).
Feramisco, J. D. et al. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl Acad. Sci. USA 102, 3242–3247 (2005).
Yang, T. et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 (2002).
Radhakrishnan, A., Goldstein, J. L., McDonald, J. G. & Brown, M. S. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8, 512–521 (2008).
Motamed, M. et al. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J. Biol. Chem. 286, 18002–18012 (2011).
Zhang, Y., Motamed, M., Seemann, J., Brown, M. S. & Goldstein, J. L. Point mutation in luminal loop 7 of Scap protein blocks interaction with loop 1 and abolishes movement to Golgi. J. Biol. Chem. 288, 14059–14067 (2013).
Jeon, T.-I. & Osborne, T. F. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 (2012).
Sever, N., Yang, T., Brown, M. S., Goldstein, J. L. & DeBose-Boyd, R. A. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 (2003).
Song, B.-L., Sever, N. & DeBose-Boyd, R. A. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 (2005).
Birsoy, K. et al. Cellular program controlling the recovery of adipose tissue mass: an in vivo imaging approach. Proc. Natl Acad. Sci. USA 105, 12985–12990 (2008).
Wrann, C. D. et al. FOSL2 promotes leptin gene expression in human and mouse adipocytes. J. Clin. Invest. 122, 1010–1021 (2012).
Clément, K. et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 392, 398–401 (1998).
Zhang, Y. et al. Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425–432 (1994). In this seminal paper, the mouse Ob gene and its human homologue LEP are identified.
Lee, G. H. et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature 379, 632–635 (1996).
Scherer, P. E., Williams, S., Fogliano, M., Baldini, G. & Lodish, H. F. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 270, 26746–26749 (1995).
Hu, E., Liang, P. & Spiegelman, B. M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 271, 10697–10703 (1996).
Shehzad, A., Iqbal, W., Shehzad, O. & Lee, Y. S. Adiponectin: regulation of its production and its role in human diseases. Hormones (Athens) 11, 8–20 (2012).
Maeda, N. et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nature Med. 8, 731–737 (2002).
Kadowaki, T. et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 116, 1784–1792 (2006).
Waki, H. & Tontonoz, P. Endocrine functions of adipose tissue. Annu. Rev. Pathol. 2, 31–56 (2007).
Takahashi, M. et al. Genomic structure and mutations in adipose-specific gene, adiponectin. Int. J. Obes. Relat. Metab. Disord. 24, 861–868 (2000).
Hara, K. et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes 51, 536–540 (2002).
Kondo, H. et al. Association of adiponectin mutation with type 2 diabetes: a candidate gene for the insulin resistance syndrome. Diabetes 51, 2325–2328 (2002).
Ibba, M. & Soll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000).
Dong, J., Qiu, H., Garcia-Barrio, M., Anderson, J. & Hinnebusch, A. G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6, 269–279 (2000).
Berlanga, J. J., Santoyo, J. & De Haro, C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem. 265, 754–762 (1999).
Scheuner, D. et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176 (2001).
Zhang, P. et al. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 (2002).
Maurin, A.-C. et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 1, 273–277 (2005).
Hao, S. et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307, 1776–1778 (2005).
Guo, F. & Cavener, D. R. The GCN2 eIF2α kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 5, 103–114 (2007).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Garami, A. et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11, 1457–1466 (2003).
Inoki, K., Li, Y., Xu, T. & Guan, K.-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003).
Tee, A. R., Manning, B. D., Roux, P. P., Cantley, L. C. & Blenis, J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 (2003).
Zhang, Y. et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nature Cell Biol. 5, 578–581 (2003).
Hara, K. et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 (1998). This paper explores the amino-acid essentiality for mTORC1 activation, and specific amino-acid requirements independent of the growth-factor-mediated regulation of activity.
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K.-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nature Cell Biol. 10, 935–945 (2008).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). References 54 and 55 report the identification of the Rag GTPases as the direct link between amino-acids levels and mTORC1, regulating mTORC1's subcellular localization.
Sancak, Y. et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Bar-Peled, L., Schweitzer, L. D., Zoncu, R. & Sabatini, D. M. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012).
Bar-Peled, L. et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013).
Panchaud, N., Péli-Gulli, M.-P. & De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42 (2013).
Tsun, Z.-Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013).
Petit, C. S., Roczniak-Ferguson, A. & Ferguson, S. M. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202, 1107–1122 (2013).
Chantranupong, L. et al. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 9, 1–8 (2014).
Peng, M., Yin, N. & Li, M. O. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 159, 122–133 (2014).
Kitamoto, K., Yoshizawa, K., Ohsumi, Y. & Anraku, Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J. Bacteriol. 170, 2683–2686 (1988).
Binda, M. et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 (2009).
Harms, E., Gochman, N. & Schneider, J. A. Lysosomal pool of free-amino acids. Biochem. Biophys. Res. Commun. 99, 830–836 (1981).
Neuhaus, E. M., Almers, W. & Soldati, T. Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Mol. Biol. Cell 13, 1390–1407 (2002).
Lee, J. H. et al. De novo somatic mutations in components of the PI(3)K–AKT3-mTOR pathway cause hemimegalencephaly. Nature Genet. 44, 941–945 (2012).
Bohn, G. et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nature Med. 13, 38–45 (2007).
Efeyan, A., Zoncu, R. & Sabatini, D. M. Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med. 18, 524–533 (2012).
Bachmanov, A. A. & Beauchamp, G. K. Taste receptor genes. Annu. Rev. Nutr. 27, 389–414 (2007).
Damak, S. et al. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science 301, 850–853 (2003).
Nelson, G. et al. An amino-acid taste receptor. Nature 416, 199–202 (2002).
Chaudhari, N. & Roper, S. D. The cell biology of taste. J. Cell Biol. 190, 285–296 (2010).
Wu, S. V. et al. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc. Natl Acad. Sci. USA 99, 2392–2397 (2002).
Wauson, E. M. et al. The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol. Cell 47, 851–862 (2012).
Printz, R. L., Magnuson, M. A. & Granner, D. K. Mammalian glucokinase. Annu. Rev. Nutr. 13, 463–496 (1993).
Nordlie, R. C., Foster, J. D. & Lange, A. J. Regulation of glucose production by the liver. Annu. Rev. Nutr. 19, 379–406 (1999).
Ogunnowo-Bada, E. O., Heeley, N., Brochard, L. & Evans, M. L. Brain glucose sensing, glucokinase and neural control of metabolism and islet function. Diabetes Obes. Metab. 16 (Suppl 1), 26–32 (2014).
Gloyn, A. L. Glucokinase (GCK) mutations in hyper- and hypoglycemia: maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemia of infancy. Hum. Mutat. 22, 353–362 (2003).
Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999).
Thorens, B. & Mueckler, M. Glucose transporters in the 21st Century. Am. J. Physiol. Endocrinol. Metab. 298, E141–E145 (2010).
Santer, R. et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nature Genet. 17, 324–326 (1997).
De Vos, A. et al. Human and rat β cells differ in glucose transporter but not in glucokinase gene expression. J. Clin. Invest. 96, 2489–2495 (1995).
Chang-Chen, K. J., Mullur, R. & Bernal-Mizrachi, E. β-Cell failure as a complication of diabetes. Rev. Endocr. Metab. Disord. 9, 329–343 (2008).
Leto, D. & Saltiel, A. R. Regulation of glucose transport by insulin: traffic control of GLUT4. Nature Rev. Mol. Cell Biol. 13, 383–396 (2012).
Hardie, D. G. AMP-activated protein kinase — an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 (2011).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev. Mol. Cell Biol. 13, 251–262 (2012).
Efeyan, A. et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013).
Zoncu, R. et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 (2011).
Zhang, F. et al. Molecular mechanism for the umami taste synergism. Proc. Natl Acad. Sci. USA 105, 20930–20934 (2008).
Nelson, G. et al. Mammalian sweet taste receptors. Cell 106, 381–390 (2001). This paper reports the identification of T1R2–T1R3 as the sweet taste receptor by means of mouse transgenesis and heterologous expression in cultured cells.
Mace, O. J., Affleck, J., Patel, N. & Kellett, G. L. Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. J. Physiol. 582, 379–392 (2007).
Dyer, J., Salmon, K. S. H., Zibrik, L. & Shirazi-Beechey, S. P. Expression of sweet taste receptors of the T1R family in the intestinal tract and enteroendocrine cells. Biochem. Soc. Trans. 33, 302–305 (2005).
Nettleton, J. A. et al. Diet soda intake and risk of incident metabolic syndrome and type 2 diabetes in the Multi-Ethnic Study of Atherosclerosis (MESA). Diabetes Care 32, 688–694 (2009).
Klionsky, D. J. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 (2000).
Egan, D. F. et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 (2011).
Kim, J., Kundu, M., Viollet, B. & Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 13, 132–141 (2011).
Mammucari, C. et al. FOXO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). This paper demonstrates the essentiality of autophagy as a crucial mechanism to mobilize internal energy stores and to adapt to the interruption of transplacental nutrient supply in neonates.
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Naito, T., Kuma, A. & Mizushima, N. Differential contribution of insulin and amino acids to the mTORC1-autophagy pathway in the liver and muscle. J. Biol. Chem. 288, 21074–21081 (2013).
Kroemer, G., Mariño, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
Tsukamoto, S. et al. Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008).
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Karsli-Uzunbas, G. et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 (2014).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
de Cabo, R., Carmona-Gutierrez, D., Bernier, M., Hall, M. N. & Madeo, F. The search for antiaging interventions: from elixirs to fasting regimens. Cell 157, 1515–1526 (2014).
Acknowledgements
D.M.S. is supported by grants from the National Institutes of Health (R01 CA129105, CA103866 and AI047389; R21 AG042876) and awards from the American Federation for Aging, Starr Foundation, Koch Institute Frontier Research Program, and the Ellison Medical Foundation. A.E. is supported by the Charles King's Trust Foundation/Simeon J. Fortin Fellowship. W.C.C. is supported by American Cancer Society – Ellison Foundation Postdoctoral Fellowship (PF-13-356-01-TBE). D.M.S. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Efeyan, A., Comb, W. & Sabatini, D. Nutrient-sensing mechanisms and pathways. Nature 517, 302–310 (2015). https://doi.org/10.1038/nature14190
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14190
This article is cited by
-
Effects of different colors of plastic-film mulching on soil temperature, yield, and metabolites in Platostoma palustre
Scientific Reports (2024)
-
Nitric oxide and ion channels mediate l-cysteine-induced inhibition of colonic smooth muscle contraction
Journal of Muscle Research and Cell Motility (2024)
-
An accurate DFT study within conformational survey of the d-form serine−alanine protected dipeptide
BMC Chemistry (2023)
-
The hunger strikes back: an epigenetic memory for autophagy
Cell Death & Differentiation (2023)
-
D-mannose induces TFE3-dependent lysosomal degradation of EGFR and inhibits the progression of NSCLC
Oncogene (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.