Abstract
Evolutionary expansion of the human neocortex underlies many of our unique mental abilities. This expansion has been attributed to the increased proliferative potential1,2 of radial glia (RG; neural stem cells) and their subventricular dispersion from the periventricular niche3,4,5 during neocortical development. Such adaptations may have evolved through gene expression changes in RG. However, whether or how RG gene expression varies between humans and other species is unknown. Here we show that the transcriptional profiles of human and mouse neocortical RG are broadly conserved during neurogenesis, yet diverge for specific signalling pathways. By analysing differential gene co-expression relationships between the species, we demonstrate that the growth factor PDGFD is specifically expressed by RG in human, but not mouse, corticogenesis. We also show that the expression domain of PDGFRβ, the cognate receptor6,7 for PDGFD, is evolutionarily divergent, with high expression in the germinal region of dorsal human neocortex but not in the mouse. Pharmacological inhibition of PDGFD–PDGFRβ signalling in slice culture prevents normal cell cycle progression of neocortical RG in human, but not mouse. Conversely, injection of recombinant PDGFD or ectopic expression of constitutively active PDGFRβ in developing mouse neocortex increases the proportion of RG and their subventricular dispersion. These findings highlight the requirement of PDGFD–PDGFRβ signalling for human neocortical development and suggest that local production of growth factors by RG supports the expanded germinal region and progenitor heterogeneity of species with large brains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Microarray data from the GCASS dataset have been deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE62064.
References
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nature Rev. Neurosci. 10, 724–735 (2009)
Lui, J. H., Hansen, D. V. & Kriegstein, A. R. Development and evolution of the human neocortex. Cell 146, 18–36 (2011)
Smart, I. H. et al. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53 (2002)
Hansen, D. V., Lui, J. H., Parker, P. R. & Kriegstein, A. R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010)
Fietz, S. A. et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature Neurosci. 13, 690–699 (2010)
Bergsten, E. et al. PDGF-D is a specific, protease-activated ligand for the PDGF β-receptor. Nature Cell Biol. 3, 512–516 (2001)
LaRochelle, W. J. et al. PDGF-D, a new protease-activated growth factor. Nature Cell Biol. 3, 517–521 (2001)
Rakic, P. Specification of cerebral cortical areas. Science 241, 170–176 (1988)
Oldham, M. C. In The OMICs: Applications in Neuroscience Vol. 1 (ed. Coppola, G. ) 85–113 (Oxford Univ. Press, 2014)
Oldham, M. C. et al. Functional organization of the transcriptome in human brain. Nature Neurosci. 11, 1271–1282 (2008)
Workman, A. D. et al. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci. 33, 7368–7383 (2013)
Fietz, S. A. et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl Acad. Sci. USA 109, 11836–11841 (2012)
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014)
Horvath, S. & Dong, J. Geometric interpretation of gene coexpression network analysis. PLOS Comput. Biol. 4, e1000117 (2008)
Oldham, M. C., Horvath, S. & Geschwind, D. H. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl Acad. Sci. USA 103, 17973–17978 (2006)
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005)
Kawaguchi, A. et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135, 3113–3124 (2008)
Pinto, L. et al. Prospective isolation of functionally distinct radial glial subtypes–lineage and transcriptome analysis. Mol. Cell. Neurosci. 38, 15–42 (2008)
Wang, Z. et al. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim. Biophys. Acta 1806, 122–130 (2010)
Diez-Roux, G. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 (2011)
Roberts, W. G. et al. Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 65, 957–966 (2005)
Magnusson, P. U. et al. Platelet-derived growth factor receptor-β constitutive activity promotes angiogenesis in vivo and in vitro. Arterioscler. Thromb. Vasc. Biol. 27, 2142–2149 (2007)
Golub, T. R., Barker, G. F., Lovett, M. & Gilliland, D. G. Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77, 307–316 (1994)
McLean, C. Y. et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471, 216–219 (2011)
Bae, B. I. et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343, 764–768 (2014)
Geschwind, D. H. & Rakic, P. Cortical evolution: judge the brain by its cover. Neuron 80, 633–647 (2013)
Johnson, M. B. et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009)
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011)
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012)
Ajioka, I., Maeda, T. & Nakajima, K. Identification of ventricular-side-enriched molecules regulated in a stage-dependent manner during cerebral cortical development. Eur. J. Neurosci. 23, 296–308 (2006)
Acknowledgements
We thank the staff at San Francisco General Hospital Women’s Options Center for their consideration in allowing access to donated human prenatal tissue. We thank J. DeYoung and the staff at the Southern California Genotyping Consortium at the University of California Los Angeles for microarray data generation. We are grateful to A. Holloway for her critical reading of the manuscript, and also thank W. Walantus, S. Wang, Y. Wang and other University of California San Francisco personnel for technical and administrative support. We thank C. Stiles and D. Rowitch for the TEL–PDGFRβ construct. This work was supported by grants from the NIH, NINDS (A.R.K.), the Bernard Osher Foundation, a California Institute for Regenerative Medicine Predoctoral Fellowship for J.H.L. (TG2-01153), a Damon Runyon Foundation Postdoctoral Fellowship for A.A.P. (DRG-2013), and the University of California San Francisco Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation (M.C.O.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the California Institute for Regenerative Medicine or any other agency of the State of California.
Author information
Authors and Affiliations
Contributions
M.C.O. conceived the GCASS strategy and J.H.L generated the GCASS data set. A.J. generated the FACS mRG data set. M.C.O. conceived, designed and performed the bioinformatic analyses. J.H.L., T.J.N. and A.A.P. designed and performed the experiments leading up to the prioritization of PDGFD as the focus of this study. T.J.N. performed the majority of the in situ hybridizations and the in vivo mouse experiments. J.H.L. performed the human and mouse slice culture experiments, as well as all of the immunostaining, imaging and image analysis in the study. M.C.O. and J.H.L. wrote the manuscript, which was edited by all the authors. M.C.O. and A.R.K provided conceptual guidance at every stage of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
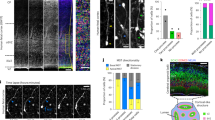
Extended Data Figure 1 Human brain dissection for GCASS and schematic for generation of FACS mRG data set.
a, Top: to generate the GCASS data set, an almost-intact prenatal human telencephalic hemisphere (GW14.5) was microdissected to separate the dorsal telencephalon from the ventral telencephalon (including medial and lateral ganglionic eminences). Bottom: the dorsal fragment was flash-frozen and serially sectioned (150 μm) for transcriptional profiling with Illumina HT-12 v4 Beadchip microarrays. Scale bars, 2.5 mm. b, To generate the FACS mRG data set, dorsal neocortices of Eomes–GFP mouse embryos were microdissected, pooled (n = 3 litters, 5–8 pooled embryos per litter), dissociated and FACS-sorted according to the gating scheme depicted to isolate RG and intermediate progenitor cells. Transcriptional profiling of the resultant populations was performed using Illumina mouseRef-8 v1.0 Beadchip microarrays.
Extended Data Figure 2 Genes comprising the six RG co-expression modules identified by GCASS are expressed in a manner consistent with the known distribution of RG in developing human neocortex.
Six candidate hRG gene co-expression modules (Fig. 1d) were superimposed on three independent gene expression data sets generated from laser-microdissected samples from prenatal human cortex: ABI.1 (ref. 13) (GW17), ABI.2 (ref. 13) (GW18), and Fietz et al. (ref. 12) (GW15–18) (as listed in Extended Data Table 1). The characteristic expression patterns of the superimposed modules were summarized by singular value decomposition; the first principal component (PC1) for each module in each data set is shown. In all cases, PC1 revealed substantially higher expression levels for these genes in germinal zones (VZ, ISVZ, and OSVZ, highlighted in red) versus non-germinal zones (IZ, SP, ICP, OCP, CP, MZ, SG). Permutation analysis indicated that the per cent variance explained (VE) by PC1 of each superimposed module was significantly greater than expected by chance (n = 10,000 permutations). CP, cortical plate; CTX, cortex; GW, gestational week; ICP, inner cortical plate; ISVZ, inner subventricular zone; IZ, intermediate zone; MZ, marginal zone; OCP, outer cortical plate; OSVZ, outer subventricular zone; SG, subpial granular layer; SP, subplate; VZ, ventricular zone.
Extended Data Figure 3 GCASS successfully predicts novel markers of neocortical hRG.
a, Genome-wide distribution of predicted GW14.5 neocortical hRG expression specificity (ZhRG). Red lines indicate predicted RG genes (validated in b, c). b, c, Immunostaining and in situ hybridization in GW14.5 human neocortex confirms RG expression specificity for novel candidate markers predicted in a (b, scale bar, 50 μm; c, scale bar, 100 μm). Analysed tissue sections were independent from the sample used for microarray analysis (Fig. 1a). ISVZ, inner subventricular zone; OSVZ, outer subventricular zone; VZ, ventricular zone.
Extended Data Figure 4 Workflow of bioinformatic procedures and experimental rationale for the entire study.
The bioinformatic component of this study sought to identify a homologous gene co-expression signature for human and mouse RG that is robust across multiple sampling strategies/technology platforms and can be normalized to facilitate comparisons within and between species. This pipeline illustrates the steps that were taken to identify, integrate and compare RG gene co-expression signatures in eight transcriptomic data sets generated from prenatal human and mouse neocortex.
Extended Data Figure 5 Genome-wide predictions of expression specificity for hRG and mRG are robust across independent data sets.
a, b, Heat maps of Spearman correlation coefficients for predicted RG expression specificity (RG PR) over 10,929 genes present in all five human data sets (a) and 10,649 genes present in all three mouse data sets (b) (as indicated in columns BE and BI in Supplementary Table 3). Data sets are denoted by the sample ages listed in Extended Data Table 1, although factors besides age also contribute to the observed correlations (for example, choice of technology platform, sample preparation strategy). E, embryonic; GW, gestational week.
Extended Data Figure 6 In situ hybridization (ISH) validates predicted presence or absence of gene expression in hRG and mRG.
a, Pink box: human in situ probes for six genes predicted to be expressed by hRG but not mRG were generated and hybridized in GW15 human neocortical tissue to validate predicted hRG expression (human scale bar, 200 μm). Blue box (Eurexpress20: http://www.eurexpress.org/ee/): in situ hybridizations for 13 genes predicted to be expressed by hRG but not mRG reveal no expression by mRG in E14.5 mouse cortex. Green box: mouse in situ probes for three genes predicted to be expressed by hRG but not mRG were generated and revealed no expression by mRG (E13.5). Positive control expression in cells other than RG are labelled in red. b, Expression patterns of genes predicted to be expressed by mRG (that is, those with the highest  values in Supplementary Table 3) are shown as further validation (blue (E14.5, Eurexpress20: http://www.eurexpress.org/ee/); orange (E14.5, GenePaint: http://www.genepaint.org); mouse scale bars, ∼500 μm). One other gene in the top 15, Cks2, is not shown, but has been validated previously30.
values in Supplementary Table 3) are shown as further validation (blue (E14.5, Eurexpress20: http://www.eurexpress.org/ee/); orange (E14.5, GenePaint: http://www.genepaint.org); mouse scale bars, ∼500 μm). One other gene in the top 15, Cks2, is not shown, but has been validated previously30.
Extended Data Figure 7 PDGFD is expressed by neocortical RG during neurogenesis in humans but not mice.
a, In situ hybridization of PDGFD in GW14.5, 16.5, 17.3 and 18.2 human neocortex demonstrates consistent expression in RG across multiple ages (scale bar, 200 μm). b, In situ hybridization of Pdgfd in E14.5 mouse (Eurexpress20: http://www.eurexpress.org/ee/) demonstrates lack of expression (scale bar, ∼500 μm). c, To demonstrate the lack of Pdgfd expression in mouse neocortex across multiple ages, a pCAG-PDGFD-IG expression plasmid was electroporated into the mouse VZ at E13.5 as an internal positive control and harvested at E14.5, E15.5 or E17.5. At E14.5 and E15.5, Pdgfd (blue signal) is seen only in the electroporated region in the ventricular zone (scale bar, 200 μm; inset scale bar, 50 μm). At E17.5, Pdgfd is in the cortical plate and not in the ventricular zone or anywhere else (scale bar, 500 μm, inset scale bar, 50 μm).
Extended Data Figure 8 Pdgfrb is strongly expressed by ventral RG and weakly expressed by lateral RG in mice.
In situ hybridization of Pdgfrb in sagittal sections through the mouse forebrain (E14.5) across a medial–lateral axis (Eurexpress20: http://www.eurexpress.org/ee/) demonstrates progenitor expression in the ventral germinal regions. This expression extends into the dorsal cortex in the lateral aspect of the brain, but is not widespread. In contrast, no progenitor expression is detected in dorsomedial cortex (scale bar, 500 μm; inset scale bar, 100 μm). Expression is also detected in the pia and vascular pericytes.
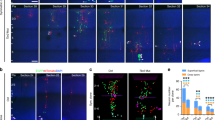
Extended Data Figure 9 Manipulation of PDGFRβ signalling in human and mouse neocortex.
a–d, Chemical blockade of PDGFRβ signalling in cultured slices of GW14.5 human neocortex impairs RG cell cycle progression. Four pharmacological inhibitors of PDGFRβ signalling were screened at different concentrations to determine their effects on RG proliferation in cultured slices of GW17.5 human neocortex (2 days). Slices were treated with BrdU for the duration of the experiment and RG proliferation was quantified as the fraction of SOX2+ cells that incorporated BrdU after treatment with inhibitor or vehicle. Statistical significance was assessed with the Wilcoxon rank sum test using the wilcox.test R function with default settings. Images are derived from ≥3 slices in each condition. Control + DMSO, n = 18; control no DMSO, n = 9; CP673451 all concentrations, n = 9; Sutent all concentrations, n = 6; imatinib (0.1 μM, 10 μM), n = 9 (100 μM), n = 6; tivozanib (1 μM, 10 μM), n = 9 (0.1 μM), n = 6. Significance indicated by: n.s. P > 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. e, Slice cultures of E13.5 mouse neocortex were treated with BrdU and DMSO (control) or a pharmacological inhibitor of PDGFRβ signalling (CP673451) for 1 or 2 days (slices from at least three independent litters). RG (SOX2+) or intermediate progenitor (TBR2+) cell proliferation was assessed as the fraction of each population that incorporated BrdU or was Ki67+ (1d: n = 10 (DMSO) versus n = 9 (CP673451); 2d: n = 11 (DMSO) versus n = 9 (CP673451)). This experiment serves as a negative control to compare with the human. f, Ectopic PDGFRβ signalling promotes RG identity in E13.5 mouse neocortex. In utero electroporation of constitutively active TEL–PDGFRβ (ref. 23) was compared with control (mouse E13.5–E15.5) and assessed for the proportion and distribution of SOX2+ RG cells or Ki67+ progenitors (out of GFP+) in the cortical wall (quantified in g: at least n = 3 slices per embryo from two independent litters, n = 15 (control); n = 18 (TEL–PDGFRβ) or (PDGFRβ(D850V)); scale bar, 50 μm). Ki67+GFP+ cell quantification following PDGFRβ(D850V)22 electroporation was performed in a similar fashion and is also shown. The spatial distributions of RG (GFP+SOX2+) in the cortical wall were assessed by quantitative image analysis (spanning ventricle to pia). The grey band delineates a 95% confidence interval for a test of equal univariate densities based on 10,000 permutations. All error bars represent mean ± s.e.m. Statistical significance for the effects of treatment was calculated by ANOVA of multiple linear regression while controlling for individual (e) and litter (f) variability (significance indicated by: n.s. P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data, Supplementary Figures 1-4, Supplementary Table 1 and additional references (see Contents page for details). (PDF 5714 kb)
Supplementary Table 1
This file shows sample characteristics of the GCASS dataset. (XLSX 61 kb)
Supplementary Table 2
The file shows genes expressed significantly higher in E14 mouse neocortical RG vs. IPC (the FACS-mRG dataset). (XLSX 53 kb)
Supplementary Table 3
This file contains a summary of predicted RG expression specificity, mean expression levels, and associated information for all genes and all datasets. (XLSX 13994 kb)
Supplementary Table 4
This file contains results of enrichment analyses for conserved and species-specific RG-expressed genes depicted in Fig. 2a, d. (XLSX 40 kb)
Rights and permissions
About this article
Cite this article
Lui, J., Nowakowski, T., Pollen, A. et al. Radial glia require PDGFD–PDGFRβ signalling in human but not mouse neocortex. Nature 515, 264–268 (2014). https://doi.org/10.1038/nature13973
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13973
This article is cited by
-
A cell fate decision map reveals abundant direct neurogenesis bypassing intermediate progenitors in the human developing neocortex
Nature Cell Biology (2024)
-
Profiling human brain vascular cells using single-cell transcriptomics and organoids
Nature Protocols (2024)
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
Developmental mechanisms underlying the evolution of human cortical circuits
Nature Reviews Neuroscience (2023)
-
The neurovascular unit in leukodystrophies: towards solving the puzzle
Fluids and Barriers of the CNS (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.