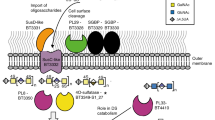
Abstract
A well-balanced human diet includes a significant intake of non-starch polysaccharides, collectively termed ‘dietary fibre’, from the cell walls of diverse fruits and vegetables1. Owing to the paucity of alimentary enzymes encoded by the human genome2, our ability to derive energy from dietary fibre depends on the saccharification and fermentation of complex carbohydrates by the massive microbial community residing in our distal gut3,4. The xyloglucans (XyGs) are a ubiquitous family of highly branched plant cell wall polysaccharides5,6 whose mechanism(s) of degradation in the human gut and consequent importance in nutrition have been unclear1,7,8. Here we demonstrate that a single, complex gene locus in Bacteroides ovatus confers XyG catabolism in this common colonic symbiont. Through targeted gene disruption, biochemical analysis of all predicted glycoside hydrolases and carbohydrate-binding proteins, and three-dimensional structural determination of the vanguard endo-xyloglucanase, we reveal the molecular mechanisms through which XyGs are hydrolysed to component monosaccharides for further metabolism. We also observe that orthologous XyG utilization loci (XyGULs) serve as genetic markers of XyG catabolism in Bacteroidetes, that XyGULs are restricted to a limited number of phylogenetically diverse strains, and that XyGULs are ubiquitous in surveyed human metagenomes. Our findings reveal that the metabolism of even highly abundant components of dietary fibre may be mediated by niche species, which has immediate fundamental and practical implications for gut symbiont population ecology in the context of human diet, nutrition and health9,10,11,12.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
McDougall, G. J., Morrison, I. M., Stewart, D. & Hillman, J. R. Plant cell walls as dietary fibre: range, structure, processing and function. J. Sci. Food Agric. 70, 133–150 (1996)
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D. & Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Rev. Microbiol. 11, 497–504 (2013)
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P. & Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306 (2012)
Koropatkin, N. M., Cameron, E. A. & Martens, E. C. How glycan metabolism shapes the human gut microbiota. Nature Rev. Microbiol. 10, 323–335 (2012)
Hoffman, M. et al. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr. Res. 340, 1826–1840 (2005)
Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11, 301–307 (2008)
Martens, E. C. et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221 (2011)
Hartemink, R., VanLaere, K. M. J., Mertens, A. K. C. & Rombouts, F. M. Fermentation of xyloglucan by intestinal bacteria. Anaerobe 2, 223–230 (1996)
Kootte, R. S. et al. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 14, 112–120 (2012)
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006)
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013)
Petrof, E. et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 1, 3 (2013)
Tasse, L. et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 20, 1605–1612 (2010)
Cummings, J. H. & Macfarlane, G. T. Role of intestinal bacteria in nutrient metabolism. Clin. Nutr. 16, 3–11 (1997)
McNeil, N. I. The contribution of the large-intestine to energy supplies in man. Am. J. Clin. Nutr. 39, 338–342 (1984)
Smith, M. I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013)
Xu, J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003)
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010)
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012)
Nelson, K. E. et al. A catalog of reference genomes from the human microbiome. Science 328, 994–999 (2010)
Yamatoya, K. & Shirakawa, M. Xyloglucan: structure, rheological properties, biological functions and enzymatic modification. Curr. Trends Polym. Sci. 8, 27–72 (2003)
Hsieh, Y. S. Y. & Harris, P. J. Xyloglucans of monocotyledons have diverse structures. Mol. Plant 2, 943–965 (2009)
Mello, L. V., Chen, X. & Rigden, D. J. Mining metagenomic data for novel domains: BACON, a new carbohydrate-binding module. FEBS Lett. 584, 2421–2426 (2010)
Cameron, E. A. et al. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J. Biol. Chem. 287, 34614–34625 (2012)
Nakjang, S., Ndeh, D. A., Wipat, A., Bolam, D. N. & Hirt, R. P. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS ONE 7, e30287 (2012)
Aspeborg, H., Coutinho, P. M., Wang, Y., Brumer, H. & Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186 (2012)
Gloster, T. M. et al. Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J. Biol. Chem. 282, 19177–19189 (2007)
Hehemann, J. H., Kelly, A. G., Pudlo, N. A., Martens, E. C. & Boraston, A. B. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Natl Acad. Sci. USA 109, 19786–19791 (2012)
Hehemann, J. H. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010)
Wegmann, U. et al. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ. Microbiol. http://dx.doi.org/10.1111/1462-2920.12217 (2013)
Koropatkin, N. M., Martens, E. C., Gordon, J. I. & Smith, T. J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 (2008)
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007)
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011)
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009)
Kurokawa, K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181 (2007)
Sonnenburg, E. D. et al. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010)
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012)
Juncker, A. S. et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 (2003)
Sundqvist, G., Stenvall, M., Berglund, H., Ottosson, J. & Brumer, H. A general, robust method for the quality control of intact proteins using LC-ESI-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 852, 188–194 (2007)
Ibatullin, F. M., Baumann, M. J., Greffe, L. & Brumer, H. Kinetic analyses of retaining endo-(xylo)glucanases from plant and microbial sources using new chromogenic xylogluco-oligosaccharide aryl glycosides. Biochemistry 47, 7762–7769 (2008)
Martinez-Fleites, C. et al. Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J. Biol. Chem. 281, 24922–24933 (2006)
Greffe, L., Bessueille, L., Bulone, V. & Brumer, H. Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology 15, 437–445 (2005)
Mopper, K. & Gindler, E. A new noncorrosive dye reagent for automatic sugar chromatography. Anal. Biochem. 56, 440–442 (1973)
McFeeters, R. F. A manual method for reducing sugar determinations with 2,2′-bicinchoninate reagent. Anal. Biochem. 103, 302–306 (1980)
Brumer, H., Sims, P. F. G. & Sinnott, M. L. Lignocellulose degradation by Phanerochaete chrysosporium: purification and characterization of the main α-galactosidase. Biochem. J. 339, 43–53 (1999)
Cartmell, A. et al. The structure and function of an arabinan-specific α-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J. Biol. Chem. 286, 15483–15495 (2011)
Miller, G. L. The use of dinitrosalicylic acid for the determination of reducing sugar. Anal. Chem. 31, 426–428 (1959)
Freelove, A. C. J., Bolam, D. N., White, P., Hazlewood, G. P. & Gilbert, H. J. A novel carbohydrate-binding protein is a component of the plant cell wall-degrading complex of Piromyces equi. J. Biol. Chem. 276, 43010–43017 (2001)
Boraston, A. B., Bolam, D. N., Gilbert, H. J. & Davies, G. J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004)
von Schantz, L. et al. Affinity maturation generates greatly improved xyloglucan-specific carbohydrate binding modules. BMC Biotechnol. 9, 92 (2009)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005)
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D 66, 470–478 (2010)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Acknowledgements
We are grateful to the following colleagues for providing materials or experimental assistance: F. Vilaplana (neutral sugar analysis of XyG preparations), G. Sundqvist (protein mass spectrometry), F. Ibatullin (aryl glycoside syntheses), S. Prexler (protein production and purification), S. Tuomivaara and W. York (provision of acetylated tomato XyG samples), the staff at the University of Michigan Germfree Laboratory (technical assistance with gnotobiotic mouse experiments) and the staff at the Diamond Light Source (provision of data collection facilities). Work in Stockholm was supported by the Mizutani Foundation for Glycoscience, The Swedish Research Council Formas (via CarboMat—the KTH Advanced Carbohydrate Materials Centre), The Swedish Research Council (Vetenskapsrådet; salary support to H.B.), and the Wallenberg Wood Science Centre (salary support to O.S. and L.S.M.). Work in Vancouver was supported by faculty funding from the Michael Smith Laboratories, University of British Columbia; the Natural Sciences and Engineering Research Council of Canada (Discovery Grant); the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund. Work in York was supported by the Biotechnology and Biological Sciences Research Council under reference BB/I014802/1. Work in Ann Arbor was supported by National Institutes of Health grants DK084214 and GM099513; T.E.R. was supported in part by the Global Probiotics Council Young Investigator Grant for Probiotics Research awarded to E.C.M.
Author information
Authors and Affiliations
Contributions
J.L. performed gene cloning, recombinant production and biochemical/enzymatic characterization for all enzymes. T.E.R. constructed B. ovatus genetic mutants and tested mutant growth phenotypes. G.R.H. performed all protein X-ray crystallography. L.S.M. performed enzyme kinetic analyses and product analyses on select enzymes and substrates (BoGH3A and 3B, BoGH5A, BoGH9A, BoGH43A and 43B). A.S.T. performed all experiments and data analysis relating to the BACON domain and carbohydrate-binding proteins. O.S. and S.K. performed initial gene cloning and production of all enzymes, and enzymatic characterization of BoGH5A and BoGH2A. N.A.P. performed growth profiling of various Bacteroides strains, including B. ovatus deletion mutants (ΔBoGH31A), on XyG oligo- and polysaccharides and analysed in vivo competition data by qPCR. K.U. analysed growth data from 292 Bacteroidetes isolates on XyG and other substrates, and assisted with metagenomic surveys. N.M.K. provided advice and assistance on XyGUL recombinant carbohydrate-binding protein production. A.L.C. and C.A.H. assisted with calorimetry and data analysis. A.G.K. assisted with comparative genomic locus identification and performed Bacteroidetes phylogenetic analysis. S.N.C. assisted with recombinant production of all enzymes and performed stability studies. E.C.M. constructed the B. ovatus Δtdk strain and the N-terminal lipidation mutant, performed corresponding cellular localization and growth studies, and performed comparative genomic analyses and metagenomic surveys. H.B., E.C.M. and G.J.D. conceived the study, directed research, analysed data, and wrote the article, including significant data analysis and writing input from J.L., T.E.R. and L.S.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Evolution of the genomic region containing the XyGUL and corresponding growth on XyG as a sole carbon source.
a, Genomic organization of 11 representative strains from 3 different species of Bacteroides. b, Growth of these strains on tamarind XyG with glucose and xylose as controls (average of n = 2 growths per strain). The observation that one B. ovatus strain (SD CMC 3f) lacks a corresponding XyGUL, as do all B. xylanisolvens (the closest cultured relative of B. ovatus), suggests that the XyGUL entered B. ovatus after it diverged. Also note that two other unrelated flanking PULs show variable presence at this locus, suggesting that it is a ‘hotspot’ for PUL evolution.
Extended Data Figure 2 Activation of putative XyGULs in Bacteroidetes isolates via growth on XyG.
a, Sentinel7 susC-like gene expression (n = 3, expression measurements from separately grown cultures relative to a minimal medium plus glucose control). b, Growth profiles of the corresponding strains in medium containing tamarind XyG with glucose and xylose as controls (average of n = 2 growths per strain). Error bars represent standard error of the mean.
Extended Data Figure 3 Cell-surface localization of BoGH5A and effect of localization on B. ovatus growth.
a, Staining of fixed wild-type and mutant B. ovatus strains. b, Western blot indicating that BoGH5A is still produced in the C1A lipidation site mutant, albeit in multiple degraded forms (residue number corresponds to the mature protein, equivalent to C33 numbered from translation start). c, Growth of wild-type (positive control), ΔBoGH5A (negative control) and BoGH5A-C1A strains on tamarind XyG. The BoGH5A-C1A mutant exhibits ∼2.6-fold slower exponential growth than the wild-type. Vertical error bars on each curve indicate the standard deviation (s.d.) of the mean (n = 3 replicates). The residual growth ability of the BoGH5A-C1A strain, despite mislocalization, is unlikely to be explained by the presence of BoGH5A enzyme accumulation in the supernatant, which was only detected by western blot for wild-type bacteria expressing BoGH5A on the cell surface. Detection of BoGH5A in panels a and b was achieved with a rabbit polyclonal antibody raised against the recombinant protein produced in E. coli (representative data from two experiments each that yielded very similar results).
Extended Data Figure 4 Non-catalytic interaction of BoGH5A variants, SusD-like Bacova_02651 and Bacova_02650 of the XyGUL with polysaccharides.
a, SDS–PAGE of recombinant proteins (representative data from at least three preparations for each protein are shown). b, Affinity gel electrophoresis (representative data from at least two gels for each experimental condition). c, Isothermal titration calorimetry (ITC); the top graph in each pair shows the raw heat during titration, whereas the bottom graph shows the integrated heats after correction. d, Association constants and thermodynamic parameters obtained from ITC data. Bovine serum albumin (BSA) was used as a non-interacting negative control protein. Other protein names were abbreviated as follows: BACON, residues Asp 37–Tyr 137 corresponding to the BACON domain of BoGH5A; Bacova_02651, SusD-like XyGUL gene product; Bacova_02650, SusE-positioned XyGUL gene product; BoGH5A E430A, full-length (Asp 37–Asn 502) catalytic nucleophile mutant of BoGH5A; CAT E430A, catalytic nucleophile mutant of the BoGH5A catalytic domain only (Ile 138–Asn 502). Reducing-sugar assays confirmed that the catalytic mutants had no detectable hydrolytic activity on XyG (data not shown), whereas an active variant (that is, E430) of CAT had a twofold higher specific activity than the full-length, wild-type BoGH5A at saturating XyG concentrations (0.5–3 mM).
Extended Data Figure 5 Abundance of Bacteroides XyGULs in human from a survey of metagenomic sequencing data from a total of 250 adult human samples.
The samples were from 211 healthy individuals, 27 with ulcerative colitis and 12 with Crohn’s disease (see Methods for references). Data sets were individually queried by BLAST using the entire XyGUL nucleotide sequence from each of the four Bacteroides species listed at the top (compare with Fig. 2) and a PUL involved in degrading the red algal polysaccharide porphyran. Each horizontal line represents the presence or absence of a hit in a single individual. The leftmost column summarizes the total XyGUL content in each person (annotated according to the colour key in the top right corner). The XyGUL frequency across all 250 samples is shown at the bottom for each condition. The graph at the far right illustrates the variation in sequencing depth for each sample/study: black lines show the average depth in megabases (Mb) for each study; the light grey line shows the depth for each individual sample.
Extended Data Figure 6 Presence of the XyGUL confers a fitness advantage to B. ovatus in the presence of dietary XyG, but only when other dietary polysaccharides are eliminated.
a, MALDI analysis of BoGH5A-digested alkaline extract from a custom mouse diet that contained a large amount of XyG from natural vegetable sources (equal amounts of cooked bell pepper, aubergine, tomato and lettuce; see Methods), indicating the presence of both solanaceous arabinogalactoxyloglucan and fucogalactoxyloglucan. b, qPCR analysis of XyGUL sentinel gene7 expression in wild-type B. ovatus grown on extracted polysaccharides from the XyG-rich custom diet, demonstrating that it significantly activates XyGUL expression over a glucose control (error bars show the s.d. of the mean of three biological replicates for both growth conditions). c, In vitro growth of wild-type and ΔXyGUL B. ovatus strains in the polysaccharide extract from the XyG-rich diet, including glucose and tamarind XyG as positive and negative control substrates, respectively. Compared with growth on tamarind XyG (middle panel), the incomplete growth defect of the ΔXyGUL mutant on the food extract (right panel) indicates that the food contains other polysaccharides that are accessible by B. ovatus. Vertical error bars on each curve indicate the s.d. of the mean of three replicates. d, In vivo competition between wild-type and ΔXyGUL B. ovatus strains in mice consuming various amounts of dietary XyG. All mice were initially fed a synthetic diet containing glucose as the sole digestible carbohydrate for 1 week and then gavaged with a 7:3 ratio of ΔXyGUL:wild type (based on independent culture optical densities, total of ∼108 viable B. ovatus) and the communities were allowed to equilibrate for 3 days. Despite the initial ratio being biased in favour of the ΔXyGUL strain, the communities equilibrated in the range 5:5–4:6, but thereafter remained stable while mice were maintained on the XyG-free diet (blue boxes in three competition plots). After community stabilization, three different dietary regimens were analysed. Left, mice were maintained on the control diet (glucose only, devoid of XyG) between days 5–37, but switched to water containing 0.25% purified XyG for days 15–37 (grey box). Middle, mice were switched to a XyG-rich, custom diet from natural food sources while simultaneously drinking water containing 0.25% purified XyG (green box). These mice were then switched to the glucose-only, XyG-free control diet while remaining on water containing 0.25% XyG (grey box). Right, mice were switched to the XyG-rich diet between days 3 and 15 but given normal water (yellow box); these mice were not continued further on any dietary regimen. Maintenance on either XyG food/XyG water (middle panel) or XyG food only (right panel) does not exert a measurable fitness pressure on the competing wild-type and ΔXyGUL strains. However, when the complex natural food polysaccharides were withheld while 0.25% XyG was maintained in water, a clear fitness pressure was observed through the significant, sequential reduction of the ΔXyGUL mutant between days 15 and 37. These data suggest that although the XyGUL confers an advantage to B. ovatus by broadening its substrate range to include XyG, the presence of alternative oligo- and polysaccharides (for example, other hemicelluloses, pectins) in a complex vegetable-based diet is nonetheless sufficient to support strains lacking this locus in vivo. Each data point is the mean abundance of the indicated strain in four separate mice and error bars represent 1 s.d. Measurements conformed to a normal distribution based on the observation that 67% of all assay values were within 1 s.d. of their respective means. Asterisks indicate statistically significant alterations (P < 0.01; Student’s t-test, one-tailed, paired) in strain abundance relative to the day 15 samples, which immediately preceded the diet switch aimed at isolating XyG as the sole exogenous polysaccharide.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-20 and Supplementary Tables 1-3. (PDF 1095 kb)
Morphing image of the two crystallographically observed conformations of the BoGH5A catalytic domain relative to the BACON domain.
The C-terminal GH5 domain (blue) is shown moving relative to the N-terminal BACON domain (green), which is fixed in the outer membrane (cartoon) via lipidation on the N-terminal cysteine residue (see Extended Data Figure ED3). (MOV 311 kb)
Superimposition of BoGH5A:XXXG with the PpXG5:GLXG complex (PDB ID 2jeq).
The active site residues of BoGH5A and the XXXG oligosaccharide are shown with green and cyan carbons, respectively. The PpXG5 active site residues are shown with orange carbons and the GLXG oligosaccharide is shown with yellow carbons. Note that the -4’ Xyl residue of the BoGH5A:XXXG occupies the corresponding position of the -2’’ Gal of the PpXG5:GXLG complex, thus indicating the likelihood of branching-dependent binding modes. This likely contributes to the marginally (four-fold) lower activity observed for XLLG-β-CNP versus XXXG-β-CNP (Extended Data Table ED1). (MOV 1589 kb)
Rights and permissions
About this article
Cite this article
Larsbrink, J., Rogers, T., Hemsworth, G. et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 (2014). https://doi.org/10.1038/nature12907
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12907
This article is cited by
-
Current models in bacterial hemicellulase-encoding gene regulation
Applied Microbiology and Biotechnology (2024)
-
Gut Microbiota Modulation and Its Implications on Neuropathic Pain: A Comprehensive Literature Review
Pain and Therapy (2024)
-
Identifying glycan consumers in human gut microbiota samples using metabolic labeling coupled with fluorescence-activated cell sorting
Nature Communications (2023)
-
Berries in Microbiome-Mediated Gastrointestinal, Metabolic, and Immune Health
Current Nutrition Reports (2023)
-
BoGH13ASus from Bacteroides ovatus represents a novel α-amylase used for Bacteroides starch breakdown in the human gut
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.