Abstract
Eukaryotic ribosomes are assembled by a complex pathway that extends from the nucleolus to the cytoplasm and is powered by many energy-consuming enzymes1,2,3. Nuclear export is a key, irreversible step in pre-ribosome maturation4,5,6,7,8, but mechanisms underlying the timely acquisition of export competence remain poorly understood. Here we show that a conserved Saccharomyces cerevisiae GTPase Nug2 (also known as Nog2, and as NGP-1, GNL2 or nucleostemin 2 in human9) has a key role in the timing of export competence. Nug2 binds the inter-subunit face of maturing, nucleoplasmic pre-60S particles, and the location clashes with the position of Nmd3, a key pre-60S export adaptor10. Nug2 and Nmd3 are not present on the same pre-60S particles, with Nug2 binding before Nmd3. Depletion of Nug2 causes premature Nmd3 binding to the pre-60S particles, whereas mutations in the G-domain of Nug2 block Nmd3 recruitment, resulting in severe 60S export defects. Two pre-60S remodelling factors, the Rea1 ATPase and its co-substrate Rsa4, are present on Nug2-associated particles, and both show synthetic lethal interactions with nug2 mutants. Release of Nug2 from pre-60S particles requires both its K+-dependent GTPase activity and the remodelling ATPase activity of Rea1. We conclude that Nug2 is a regulatory GTPase that monitors pre-60S maturation, with release from its placeholder site linked to recruitment of the nuclear export machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Strunk, B. S. & Karbstein, K. Powering through ribosome assembly. RNA 15, 2083–2104 (2009)
Staley, J. P. & Woolford, J. L., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21, 109–118 (2009)
Granneman, S. & Baserga, S. J. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296, 43–50 (2004)
Lafontaine, D. L. J. A. ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem. Sci. 35, 267–277 (2010)
Warner, J. R. & McIntosh, K. B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 (2009)
Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell 136, 763–776 (2009)
Zemp, I. & Kutay, U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581, 2783–2793 (2007)
Dez, C., Houseley, J. & Tollervey, D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae . EMBO J. 25, 1534–1546 (2006)
Tsai, R. Y. & Meng, L. Nucleostemin: a latecomer with new tricks. Int. J. Biochem. Cell Biol. 41, 2122–2124 (2009)
Sengupta, J. et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 189, 1079–1086 (2010)
Matsuo, Y. et al. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis . J. Biol. Chem. 281, 8110–8117 (2006)
Granneman, S., Kudla, G., Petfalski, E. & Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl Acad. Sci. USA 106, 9613–9618 (2009)
Ben-Shem, A., Jenner, L., Yusupova, G. & Yusupov, M. Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209 (2010)
Gadal, O. et al. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21, 3405–3415 (2001)
Ho, J. H., Kallstrom, G. & Johnson, A. W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151, 1057–1066 (2000)
Bourne, H. R., Sanders, D. A. & McCormick, F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 (1991)
Hedges, J., West, M. & Johnson, A. W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24, 567–579 (2005)
Lapik, Y. R., Misra, J. M., Lau, L. F. & Pestov, D. G. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol. Cell. Biol. 27, 7735–7744 (2007)
Amlacher, S. et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 (2011)
Ash, M. R., Maher, M. J., Mitchell Guss, J. & Jormakka, M. The cation-dependent G-proteins: in a class of their own. FEBS Lett. 586, 2218–2224 (2012)
Ulbrich, C. et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138, 911–922 (2009)
Baßler, J. et al. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell 38, 712–721 (2010)
Kikkawa, S. et al. Conversion of GDP into GTP by nucleoside diphosphate kinase on the GTP-binding proteins. J. Biol. Chem. 265, 21536–21540 (1990)
Wertheimer, A. M. & Kaulenas, M. S. GDP kinase activity associated with salt-washed ribosomes. Biochem. Biophys. Res. Commun. 78, 565–571 (1977)
Hung, N. J. & Johnson, A. W. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae . Mol. Cell. Biol. 26, 3718–3727 (2006)
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods 6, 917–922 (2009)
Dembowski, J. A., Kuo, B. & Woolford, J. L., Jr Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res. 41, 7889–7904 (2013)
Chennupati, V. et al. Signals and pathways regulating nucleolar retention of novel putative nucleolar GTPase NGP-1(GNL-2). Biochemistry 50, 4521–4536 (2011)
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14, 953–961 (1998)
Granneman, S., Petfalski, E. & Tollervey, D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 30, 4006–4019 (2011)
Ferreira-Cerca, S. et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nature Struct. Mol. Biol. 19, 1316–1323 (2012)
Bradatsch, B. et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nature Struct. Mol. Biol. 19, 1234–1241 (2012)
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 17, 1030–1032 (1999)
Baßler, J. et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8, 517–529 (2001)
Hurt, E. et al. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144, 389–401 (1999)
SenGupta, D. J. et al. A three-hybrid system to detect RNA-protein interactions in vivo . Proc. Natl Acad. Sci. USA 93, 8496–8501 (1996)
Saveanu, C. et al. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23, 4449–4460 (2003)
de la Cruz, J., Sanz-Martinez, E. & Remacha, M. The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits. Nucleic Acids Res. 33, 5728–5739 (2005)
Gwizdek, C. et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl Acad. Sci. USA 103, 16376–16381 (2006)
Frey, S., Pool, M. & Seedorf, M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 276, 15905–15912 (2001)
Vilardell, J. & Warner, J. R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17, 1959–1965 (1997)
Lebreton, A., Saveanu, C., Decourty, L., Jacquier, A. & Fromont-Racine, M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 281, 27099–27108 (2006)
Bussiere, C., Hashem, Y., Arora, S., Frank, J. & Johnson, A. W. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 197, 747–759 (2012)
Lebreton, A. et al. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173, 349–360 (2006)
Thomas, B. J. & Rothstein, R. Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 (1989)
Baßler, J., Kallas, M. & Hurt, E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 281, 24737–24744 (2006)
Acknowledgements
We thank M. Remacha, M. Fromont- Racine, A. W. Johnson, C. Dargemont, M. Seedorf and J. Warner for antibodies. We thank the GenePool at the University of Edinburgh for performing the MiSeq sequencing, and E. Petfalski for performing the initial crosslinking experiments. We thank E. Thomson for careful reading the manuscript. This work was supported by a postdoctoral fellowship from Alexander von Humboldt Foundation to Y.M., and by the Wellcome Trust to S.G. and D.T. (077248), and by grants from the Deutsche Forschungsgemeinschaft to E.H. (DFG Hu363/10-4).
Author information
Authors and Affiliations
Contributions
Experiments were designed and the data were interpreted by Y.M. and E.H.; all experiments except CRAC analysis were performed by Y.M.; CRAC experiments and data analyses were performed by S.G. in collaboration with D.T.; M.T. constructed rea1 mutants and performed the in vitro release assay of rea1 mutants, the ctNug2 complementation assay and the immunodepletion assay; R.-G.M. developed the methods of the in vitro assay for nucleotide binding and GTPase activity measurement; the manuscript was written by Y.M. and E.H.; all authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Multiple sequence alignment of various Nug2 orthologues.
Multiple sequence alignment of YlqF (bacterial homologue of Nug2; Bacillus cereus), ctNug2, DmNug2 (Drosophila melanogaster), DrNug2 (Danio rerio), HsNug2 (Homo sapiens), KlNug2 (Kluyveromyces lactis), MmNug2 (Mus musculus), ScNug2 (S. cerevisiae), SpNug2 (S. pombe), XlNug2 (Xenopus laevis) and YlNug2 (Yarrowia lipolytica), using T-Coffee multiple sequence alignment (http://www.ebi.ac.uk/Tools/msa/tcoffee) and Jalview. Indicated above the alignment are the different Nug2 domains including the N, G, and C domains and the C-terminal extension. Moreover, the DAR, G1, G3, G4 motifs, point mutation sites (in red) and truncated site of ctNUG2-510 amino acids (truncation of the non-conserved C-terminal extension; red line) are indicated.
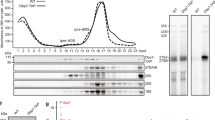
Extended Data Figure 2 Nug2 and Nmd3 are not found on the same pre-60S particles.
Indicated different TAP-tagged bait proteins were affinity purified from yeast wild-type cells. The final eluates were analysed by SDS–PAGE and Coomassie staining (top), and by western blotting using the indicated antibody (bottom). Asterisks mark the position of each bait protein. Rea1 has been identified by mass spectrometry. All affinity purifications and western analyses were performed at least twice, yielding highly reproducible data sets.
Extended Data Figure 3 ctNug2 can complement the lethal phenotype of a nug2Δ null mutant.
Serial dilutions of the yeast Nug2 shuffle strain (MAT a , ade2, ade3, his3, ura3, leu2, trp1, nug2::kanMX4, pHT4467-NUG2) transformed with empty plasmid, yeast ScNUG2, ctNUG2 or ctNUG2-510 (truncation of the non-conserved C-terminal extension; see Extended Data Fig. 1) under the control of the constitutive ADH1 promoter in a single-copy-number (YCplac111) or multi-copy-number (pRS425) plasmid (see Supplementary Table 2) were spotted on SDC−Leu (loading control) and SDC plates containing 5-FOA at indicated temperatures for 6 days. Note that ctNug2 only partially complements the nug2 null mutant.
Extended Data Figure 4 Mutations in ATP-binding or MIDAS domain of Rea1 inhibit the release of Rsa4 and Nug2 from the pre-60S particle.
a, b, Wild-type REA1 and the rea1 mutants mapping in the ATP-binding site of the AAA2 domain (Lys659Ala) or in the MIDAS domain (DAA)21 were N-terminally tagged with eGFP and expressed in a REA1 shuffle strain (a) or overexpressed under the control of the inducible GAL1-10 promoter in REA1 wild-type strain DS1-2b (b). Transformants were spotted in tenfold serial dilution steps on the indicated plates and incubated at 30 °C for 3 days. Both of the rea1 mutant alleles do not complement the rea1 null strain (a, SDC + 5-FOA) and cause a dominant-negative phenotype after overexpression by replacing endogenous Rea1 (b, galactose). c, Overnight pre-cultures were grown in SRC−Leu to prevent plasmid loss, followed by shifting cells (A600 nm = 0.75) to galactose medium (YPG) for 7 h. Rix1 particles, which were affinity purified from a Rix1–TAP, RpL3–Flag strain containing either endogenous wild-type or overexpressed wild-type eGFP–Rea1, eGFP–Rea1(DAA) and eGFP–Rea1(Lys659Ala), were incubated with or without 4 mM ATP in KCl buffer, before the different in vitro matured pre-60S particles were re-isolated by affinity-purification via the RpL3–Flag on Flag beads. Subsequently, the in vitro matured pre-60S particles (eluates) were analysed by SDS–PAGE and Coomassie staining. Relevant bands are indicated on the right. Note that in the case of the rea1 mutants, the release of Nug2, Rsa4 but also Rea1 and the Rix1 complex is significantly inhibited. All in vitro assays were performed at least twice, yielding highly reproducible data sets.
Extended Data Figure 5 Nug2 depletion assay using the auxin-inducible degron system.
a, Growth of Nug2 auxin degron strains (sAid–Nug2–sAid) in the PADH-OsTIR1 background on YPD plates with or without 500 μM auxin (IAA). The cell growth of sAid–Nug2–sAid strain was inhibited by the addition of auxin. b, Western blotting of sAid–Nug2–sAid after auxin treatment. The depletion of sAid–Nug2–sAid occurred within about 30 min of auxin addition.
Rights and permissions
About this article
Cite this article
Matsuo, Y., Granneman, S., Thoms, M. et al. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505, 112–116 (2014). https://doi.org/10.1038/nature12731
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12731
This article is cited by
-
A single 2′-O-methylation of ribosomal RNA gates assembly of a functional ribosome
Nature Structural & Molecular Biology (2023)
-
Comparative transcriptome analysis reveals the resistance regulation mechanism and fungicidal activity of the fungicide phenamacril in Fusarium oxysporum
Scientific Reports (2022)
-
Puf6 primes 60S pre-ribosome nuclear export at low temperature
Nature Communications (2021)
-
Arabidopsis REI-LIKE proteins activate ribosome biogenesis during cold acclimation
Scientific Reports (2021)
-
Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin–ligand-type complexes
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.