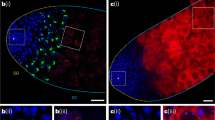
Abstract
A paramutation is an epigenetic interaction between two alleles of a locus, through which one allele induces a heritable modification in the other allele without modifying the DNA sequence1,2. The paramutated allele itself becomes paramutagenic, that is, capable of epigenetically converting a new paramutable allele. Here we describe a case of paramutation in animals showing long-term transmission over generations. We previously characterized a homology-dependent silencing mechanism referred to as the trans-silencing effect (TSE), involved in P-transposable-element repression in the germ line3,4,5. We now show that clusters of P-element-derived transgenes that induce strong TSE6,7 can convert other homologous transgene clusters incapable of TSE into strong silencers, which transmit the acquired silencing capacity through 50 generations. The paramutation occurs without any need for chromosome pairing between the paramutagenic and the paramutated loci, and is mediated by maternal inheritance of cytoplasm carrying Piwi-interacting RNAs (piRNAs) homologous to the transgenes. The repression capacity of the paramutated locus is abolished by a loss-of-function mutation of the aubergine gene involved in piRNA biogenesis, but not by a loss-of-function mutation of the Dicer-2 gene involved in siRNA production. The paramutated cluster, previously producing barely detectable levels of piRNAs, is converted into a stable, strong piRNA-producing locus by the paramutation and becomes fully paramutagenic itself. Our work provides a genetic model for the emergence of piRNA loci, as well as for RNA-mediated trans-generational repression of transposable elements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brink, R. A. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41, 872–889 (1956)
Coe, E. H., Jr A regular and continuing conversion-type phenomenon at the B locus in maize. Proc. Natl Acad. Sci. USA 45, 828–832 (1959)
Roche, S. E. & Rio, D. C. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics 149, 1839–1855 (1998)
Josse, T. et al. Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS ONE 3, e3249 (2008)
Josse, T. et al. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 3, 1633–1643 (2007)
Dorer, D. R. & Henikoff, S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics 147, 1181–1190 (1997)
Ronsseray, S., Boivin, A. & Anxolabehere, D. P-element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics 159, 1631–1642 (2001)
Chandler, V. L. Paramutation: from maize to mice. Cell 128, 641–645 (2007)
Hollick, J. B., Patterson, G. I., Coe, E. H., Cone, K. C. & Chandler, V. L. Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141, 709–719 (1995)
Pilu, R. et al. A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity 102, 236–245 (2009)
Sidorenko, L. V. & Peterson, T. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell 13, 319–335 (2001)
Stam, M. Paramutation: a heritable change in gene expression by allelic interactions in trans. Molecular Plant 2, 578–588 (2009)
Alleman, M. et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442, 295–298 (2006)
Dorweiler, J. E. et al. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12, 2101–2118 (2000)
Arteaga-Vazquez, M. et al. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc. Natl Acad. Sci. USA 107, 12986–12991 (2010)
Stam, M., Belele, C., Dorweiler, J. E. & Chandler, V. L. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918 (2002)
Rassoulzadegan, M. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006)
Grandjean, V. et al. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development 136, 3647–3655 (2009)
Kidwell, M. G., Kidwell, J. F. & Sved, J. A. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics 86, 813–833 (1977)
Engels, W. R. in P Elements in Drosophila (eds Berg, D. E., & Howe, M. M. ) (American Society for Microbiology, 1989)
Ronsseray, S., Lehmann, M., Nouaud, D. & Anxolabehere, D. The regulatory properties of autonomous subtelomeric P elements are sensitive to a suppressor of variegation in Drosophila melanogaster. Genetics 143, 1663–1674 (1996)
Marin, L. et al. P-element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics 155, 1841–1854 (2000)
Stuart, J. R. et al. Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics 162, 1641–1654 (2002)
Poyhonen, M. et al. Homology-dependent silencing by an exogenous sequence in the Drosophila germline. G3 (Bethesda) 2, 331–338 (2012)
Todeschini, A. L., Teysset, L., Delmarre, V. & Ronsseray, S. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS ONE 5, e11032 (2010)
Dorer, D. R. & Henikoff, S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77, 993–1002 (1994)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007)
Czech, B. et al. An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 (2008)
Muerdter, F. et al. Production of artificial piRNAs in flies and mice. RNA 18, 42–52 (2011)
Lemaitre, B., Ronsseray, S. & Coen, D. Maternal repression of the P element promoter in the germline of Drosophila melanogaster: a model for the P cytotype. Genetics 135, 149–160 (1993)
O’Kane, C. J. & Gehring, W. J. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl Acad. Sci. USA 84, 9123–9127 (1987)
Bier, E. et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3, 1273–1287 (1989)
Schupbach, T. & Wieschaus, E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136 (1991)
Wilson, J. E., Connell, J. E., Schlenker, J. D. & Macdonald, P. M. Novel genetic screen for genes involved in posterior body patterning in Drosophila. Dev. Genet. 19, 199–209 (1996)
Harris, A. N. & Macdonald, P. M. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128, 2823–2832 (2001)
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Acknowledgements
We thank O. Sismeiro, J.-Y. Copée, E. Mouchel-Vielh, V. Ribeiro, C. Pappatico and P. Graça for technical assistance, D. Dorer, S. Henikoff and the Bloomington Stock Center for providing stocks, and flybase.org for providing databases. We thank T. Josse for preliminary experiments. We thank J.-R. Huynh, V. Colot, N. Randsholt, A.-M. Pret, C. Carré and F. Peronnet for critical reading of the manuscript. S.R. thanks D. Anxolabéhère and M. Lehmann for previous help. This work was supported by fellowships from the Ministère de l’Enseignement Supérieur et de la Recherche to A.d.V. and C.H., from the Fondation pour la Recherche Médicale to A.d.V., from the Association Nationale de la Recherche (ANR) to A.-L.B., and by grants from the Association pour la Recherche contre le Cancer to S.R. and from the ANR (project “Nuclear endosiRNAs”) to C.A.
Author information
Authors and Affiliations
Contributions
Genetic experiments were conceived by A.d.V., A.B. and S.R., and performed by A.d.V., A.B., C.H., V.D., L.T. and S.R. L.T. conceived and performed molecular mapping of the clusters and Southern blot analysis. Deep-sequencing analysis was conceived by A.d.V., A.-L.B., S.R. and C.A., and performed by A.d.V. and A.-L.B. Bioinformatic analysis was conceived and performed by C.A. RT–qPCR was conceived and performed by A.B. S.R., A.d.V., A.B. and C.A. wrote the paper and all authors discussed the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-9, Supplementary Tables 1-4, a Supplementary Discussion and Supplementary References. (PDF 1822 kb)
Rights and permissions
About this article
Cite this article
de Vanssay, A., Bougé, AL., Boivin, A. et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490, 112–115 (2012). https://doi.org/10.1038/nature11416
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11416
This article is cited by
-
The composition of piRNA clusters in Drosophila melanogaster deviates from expectations under the trap model
BMC Biology (2023)
-
Modeling early germline immunization after horizontal transfer of transposable elements reveals internal piRNA cluster heterogeneity
BMC Biology (2023)
-
Cisplatin exposure alters tRNA-derived small RNAs but does not affect epimutations in C. elegans
BMC Biology (2023)
-
A maternally programmed intergenerational mechanism enables male offspring to make piRNAs from Y-linked precursor RNAs in Drosophila
Nature Cell Biology (2023)
-
Small RNAs mediate transgenerational inheritance of genome-wide trans-acting epialleles in maize
Genome Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.