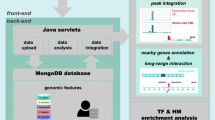
Abstract
Systematic annotation of gene regulatory elements is a major challenge in genome science. Direct mapping of chromatin modification marks and transcriptional factor binding sites genome-wide1,2 has successfully identified specific subtypes of regulatory elements3. In Drosophila several pioneering studies have provided genome-wide identification of Polycomb response elements4, chromatin states5, transcription factor binding sites6,7,8,9, RNA polymerase II regulation8 and insulator elements10; however, comprehensive annotation of the regulatory genome remains a significant challenge. Here we describe results from the modENCODE cis-regulatory annotation project. We produced a map of the Drosophila melanogaster regulatory genome on the basis of more than 300 chromatin immunoprecipitation data sets for eight chromatin features, five histone deacetylases and thirty-eight site-specific transcription factors at different stages of development. Using these data we inferred more than 20,000 candidate regulatory elements and validated a subset of predictions for promoters, enhancers and insulators in vivo. We identified also nearly 2,000 genomic regions of dense transcription factor binding associated with chromatin activity and accessibility. We discovered hundreds of new transcription factor co-binding relationships and defined a transcription factor network with over 800 potential regulatory relationships.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000)
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007)
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009)
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B. & Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 (2007)
Filion, G. J. et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 (2010)
Li, X. Y. et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27 (2008)
MacArthur, S. et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80 (2009)
Zeitlinger, J. et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature Genet. 39, 1512–1516 (2007)
Zinzen, R. P., Girardot, C., Gagneur, J., Braun, M. & Furlong, E. E. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009)
Nègre, N. et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6, e1000814 (2010)
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007)
Schotta, G. et al. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21, 1121–1131 (2002)
Agger, K., Christensen, J., Cloos, P. A. & Helin, K. The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 18, 159–168 (2008)
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009)
Kwong, C. et al. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 4, e1000178 (2008)
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 41, 376–381 (2009)
Hoskins, R. A. et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster . Genome Res. 21, 182–192 (2011)
Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010)
Moorman, C. et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster . Proc. Natl Acad. Sci. USA 103, 12027–12032 (2006)
Henikoff, S., Henikoff, J. G., Sakai, A., Loeb, G. B. & Ahmad, K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 19, 460–469 (2009)
Gebelein, B., McKay, D. J. & Mann, R. S. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 431, 653–659 (2004)
Nolo, R., Abbott, L. A. & Bellen, H. J. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila . Cell 102, 349–362 (2000)
Haecker, A. et al. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biol. 5, e145 (2007)
Rivera-Pomar, R., Lu, X., Perrimon, N., Taubert, H. & Jackle, H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature 376, 253–256 (1995)
Zraly, C. B. et al. SNR1 is an essential subunit in a subset of Drosophila brm complexes, targeting specific functions during development. Dev. Biol. 253, 291–308 (2003)
Truman, J. W. Metamorphosis of the central nervous system of Drosophila . J. Neurobiol. 21, 1072–1084 (1990)
Parrish, J. Z., Kim, M. D., Jan, L. Y. & Jan, Y. N. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 20, 820–835 (2006)
Acknowledgements
This work was supported by U01HG004264 from the National Human Genome Research Institute to K.P.W. and also funded by the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Human Genome Research Institute (NHGRI) or the National Institutes of Health (NIH). C.D.B. is supported by a Lilly-Life Sciences Research Foundation fellowship. C.A.B. is supported by a NIH NRSA postdoctoral fellowship. R.P.A. is in part supported by an Isaac Newton Trust award to R.W. P.L. was supported by a grant from the Department of Energy Computational Sciences Graduate Fellowship (DOE CSGF). M.E.L. and D.M.M. work was supported by NHGRI grant U01 HG004279. We thank the Functional Genomics Facility at the University of Chicago and the High-Throughput Genome Analysis Core at Argonne National Laboratory for processing of microarrays and of Illumina sequence. We thank T.-R. Li, J. D. Lambert, S. Rifkin, T. Herreman, C. Mason, L. Sun and Z. Gauhar for producing the developmental expression microarray data. We also thank the many members of the Drosophila community who contributed to this work by providing reagents. A complete list of community participants is included in the Supplementary Methods.
Author information
Authors and Affiliations
Contributions
N.N., L.S. and K.P.W. designed and produced modENCODE antibodies; N.N., Z.L., H.I., R.F.S., M.B.D., C.A.M., J.Z., S.S. and M.D. performed the ChIP-chip and ChIP-seq experiments; R.F.S., K.V., H.B. and A.V. produced the GFP-tagged transcription factor Drosophila lines; S.W.M., H.I., L.H. and R.P.A. performed the validation experiments of promoters, enhancers and insulators; N.N., P.K.S., N.A.B., A.J.G., D.H. and R.L.G. performed the primary analysis and organized the data of ChIP-chip and ChIP-seq experiments; N.N., C.D.B., L.M., C.A.B., U.W., P.K., M.L.E., P.L., R.S., J.C., C.C., P.K.S., D.M.M. and M.G. analysed the data; M.M. contributed to reagents; N.N., C.D.B., L.M., C.A.B., S.W.M., R.P.A., R.W., S.R., B.R., M.G., J.W.P., M.K. and K.P.W. wrote the paper; H.B., R.W., S.R. (silencer/insulator analysis), R.L.G. (informatics), B.R. (chromatin data and promoter validation), J.W.P. (enhancer/promoter validation), M.K. (data analysis) and K.P.W. (project director) supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, additional references, Supplementary Tables 1, 2, 5, 9, 16 and legends for Tables 1-16 (see separate files for tables 3, 6-8 and 10-15) and Supplementary Figures and legends 1-28. (PDF 26217 kb)
Supplementary Table 3
Promoter validation results - see Supplementary Information file for full legend. (XLS 141 kb)
Supplementary Table 6
TSS class annotation at FDR 0.05 - see Supplementary Information file for full legend. (TXT 9092 kb)
Supplementary Table 7
TSS class annotation at FDR 0.1 - see Supplementary Information file for full legend (TXT 9092 kb)
Supplementary Table 8
Novel promoter prediction based on co-occurence of H3K4me3, PolII and RNA in embryos - see Supplementary Information file for full legend. (TXT 202 kb)
Supplementary Table 10
Insulators Class I - see Supplementary Information file for full legend. (TXT 106 kb)
Supplementary Table 11
Insulators Class II - see Supplementary Information file for full legend. (TXT 65 kb)
Supplementary Table 12
HDAC associated PREs - see Supplementary Information file for full legend. (TXT 16 kb)
Supplementary Table 13
CBP embryo only enhancer predictions - see Supplementary Information file for full legend. (TXT 2 kb)
Supplementary Table 14
TF driven clustering of CBP bound regions - see Supplementary Information file for full legend. (XLS 1278 kb)
Supplementary Table 15
Enrichment of CBP developmental stages within CBP clusters - see Supplementary Information file for full legend. (XLS 32 kb)
Rights and permissions
About this article
Cite this article
Nègre, N., Brown, C., Ma, L. et al. A cis-regulatory map of the Drosophila genome. Nature 471, 527–531 (2011). https://doi.org/10.1038/nature09990
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09990
This article is cited by
-
Identification of transcription factor high accumulation DNA zones
BMC Bioinformatics (2023)
-
CRISPR/Cas9 and FLP-FRT mediated regulatory dissection of the BX-C of Drosophila melanogaster
Chromosome Research (2023)
-
ASC proneural factors are necessary for chromatin remodeling during neuroectodermal to neuroblast fate transition to ensure the timely initiation of the neural stem cell program
BMC Biology (2022)
-
Histone deacetylation primes self-propagation of heterochromatin domains to promote epigenetic inheritance
Nature Structural & Molecular Biology (2022)
-
Transcription factor paralogs orchestrate alternative gene regulatory networks by context-dependent cooperation with multiple cofactors
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.