Abstract
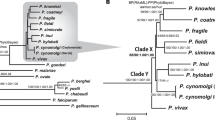
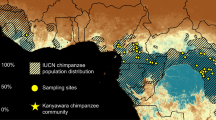
Plasmodium falciparum is the most prevalent and lethal of the malaria parasites infecting humans, yet the origin and evolutionary history of this important pathogen remain controversial. Here we develop a single-genome amplification strategy to identify and characterize Plasmodium spp. DNA sequences in faecal samples from wild-living apes. Among nearly 3,000 specimens collected from field sites throughout central Africa, we found Plasmodium infection in chimpanzees (Pan troglodytes) and western gorillas (Gorilla gorilla), but not in eastern gorillas (Gorilla beringei) or bonobos (Pan paniscus). Ape plasmodial infections were highly prevalent, widely distributed and almost always made up of mixed parasite species. Analysis of more than 1,100 mitochondrial, apicoplast and nuclear gene sequences from chimpanzees and gorillas revealed that 99% grouped within one of six host-specific lineages representing distinct Plasmodium species within the subgenus Laverania. One of these from western gorillas comprised parasites that were nearly identical to P. falciparum. In phylogenetic analyses of full-length mitochondrial sequences, human P. falciparum formed a monophyletic lineage within the gorilla parasite radiation. These findings indicate that P. falciparum is of gorilla origin and not of chimpanzee, bonobo or ancient human origin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
SGA-derived Plasmodium nucleotide sequences have been deposited in GenBank under accession numbers HM234976–HM235117 and HM237301 (cytb), HM235118–HM235143 (ldh), HM235144–HM235170 (clpC), HM235171–HM235268 (mtDNA-3.3 kb) and HM235269–HM235404 (mtDNA-3.4 kb) (also see Supplementary Table 6).
References
Greenwood, B. M., Bojang, K., Whitty, C. J. & Targett, G. A. Malaria. Lancet 365, 1487–1498 (2005)
Snow, R. W., Guerra, C. A., Noor, A. M., Myint, H. Y. & Hay, S. I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434, 214–217 (2005)
Carter, R. & Mendis, K. N. Evolutionary and historical aspects of the burden of malaria. Clin. Microbiol. Rev. 15, 564–594 (2002)
Kappe, S. H., Vaughan, A. M., Boddey, J. A. & Cowman, A. F. That was then but this is now; malaria research in the time of an eradication agenda. Science 328, 862–866 (2010)
Escalante, A. A. & Ayala, F. J. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl Acad. Sci. USA 91, 11373–11377 (1994)
Escalante, A. A., Barrio, E. & Ayala, F. J. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol. Biol. Evol. 12, 616–626 (1995)
Jeffares, D. C. et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum . Nature Genet. 39, 120–125 (2006)
Rich, S. M., Licht, M. C., Hudson, R. R. & Ayala, F. J. Malaria’s Eve: evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum . Proc. Natl Acad. Sci. USA 95, 4425–4430 (1998)
Rich, S. M. et al. The origin of malignant malaria. Proc. Natl Acad. Sci. USA 106, 14902–14907 (2009)
Prugnolle, F. et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum . Proc. Natl Acad. Sci. USA 107, 1458–1463 (2010)
Krief, S. et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from bonobos. PLoS Pathogens 6, e1000765 (2010)
Duval, L. et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc. Natl Acad. Sci. USA 107, 10561–10566 (2010)
Liu, W. et al. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathogens 4, e1000097 (2008)
Keele, B. F. et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526 (2006)
Neel, C. et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J. Virol. 84, 1464–1476 (2010)
Van Heuverswyn, F. et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368, 155–171 (2007)
Nwakanma, D. C. et al. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J. Infect. Dis. 199, 1567–1574 (2009)
Bray, R. S. Studies on malaria in chimpanzees. VI. Laverania falciparum . Am. J. Trop. Med. Hyg. 7, 20–24 (1958)
Simmonds, P. et al. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 64, 864–872 (1990)
Palmer, S. et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43, 406–413 (2005)
Salazar-Gonzalez, J. F. et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82, 3952–3970 (2008)
Keele, B. F. et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl Acad. Sci. USA 105, 7552–7557 (2008)
Keele, B. F. et al. Low dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206, 1117–1134 (2009)
Su, X.-Z., Mu, J. & Joy, D. A. The “Malaria’s Eve” hypothesis and the debate concerning the origin of the human malaria parasite Plasmodium falciparum . Microbes Infect. 5, 891–896 (2003)
Coluzzi, M. The clay feet of the malaria giant and its African roots: hypotheses and inferences about origin, spread and control of Plasmodium falciparum . Parassitologia 41, 277–283 (1999)
Martin, M. J., Rayner, J. C., Gagneux, P., Barnwell, J. W. & Varki, A. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl Acad. Sci. USA 102, 12819–12824 (2005)
Ollomo, B. et al. A new malaria agent in African hominids. PLoS Pathogens 5, e1000446 (2009)
Duval, L. et al. Chimpanzee malaria parasite related to Plasmodium ovale in Africa. PLoS ONE 4, e5520 (2009)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Guindon, S., Delsuc, F., Dufayard, J. F. & Gascuel, O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137 (2009)
Li, Y. et al. in Proc. 17th Conf. Retroviruses Opportunistic Infections abstr. 440, 〈http://www.retroconference.org/2010/PDFs/440.pdf〉 (2010)
Talman, A. M. et al. Evaluation of the intra- and inter-specific genetic variability of Plasmodium lactate dehydrogenase. Malar. J. 6, 140 (2007)
Brown, W. M. et al. Comparative structural analysis and kinetic properties of lactate dehydrogenases from the four species of human malarial parasites. Biochemistry 43, 6219–6229 (2004)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Posada, D. & Buckley, T. R. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 (2004)
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008)
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985)
Acknowledgements
We thank C. Neel, S. Loul, A. Mebanga, B. Yangda and F. Liegeois for field work in Cameroon; the Cameroonian Ministries of Health, Forestry and Wildlife, and Research for permission to collect samples in Cameroon; the Water and Forest Ministry for permission to collect samples in the Central African Republic; the Ministries of Science and Technology and Forest Economy for permission to collect samples in the Republic of the Congo; the Ministry of Scientific Research and Technology and the Department of Ecology and Management of Plant and Animal Resources of the University of Kisangani for permission to collect samples in the Democratic Republic of the Congo; M. Ndunda, S. Coxe, A. Lokasola, A. Todd and the staff of the World Wildlife Fund in the Central African Republic for logistical support; R. Carter for helpful discussions; M. Salazar, Y. Chen and B. Cochran for technical assistance; and J. White for artwork and manuscript preparation. This work was supported by grants from the National Institutes of Health (R01 AI50529, R01 AI58715, U19 AI 067854, R03 AI074778, T32 GM008111, T32 AI007245, P30 AI 27767), the Bill & Melinda Gates Foundation (37874), the National Science Foundation (0755823), the Agence Nationale de Recherche sur le Sida (12152/12182), the Great Ape Conservation Fund of the US Fish and Wildlife Service, the Arthur L. Greene Fund, the Wallace Global Fund, the Bristol Myers Freedom to Discover Program and the Wellcome Trust. R.S.R. was supported by a Howard Hughes Medical Institute Med-into-Grad Fellowship.
Author information
Authors and Affiliations
Contributions
All authors contributed to the acquisition, analysis and interpretation of the data; W.L., M.P., J.C.R., P.M.S. and B.H.H. initiated and designed the study; W.L., Y.L. and J.D.R. performed non-invasive Plasmodium testing and SGA analyses; B.F.K, R.S.R and J.D.R. performed microsatellite analyses; P.M.S. calculated Plasmodium prevalence rates; G.H.L. and P.M.S performed phylogenetic analyses; J.-B.N.N., C.M.S., D.B.M., S.L., M.K.G., P.J.K., P.D.W., E.D., E.M.-N., A.V.G. and M.N.M. conducted and supervised all fieldwork; and W.L., G.M.S., M.P., P.M.S., J.C.R. and B.H.H. coordinated the contributions of all authors and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 with legends and Supplementary Tables 1-7. (PDF 2047 kb)
Rights and permissions
About this article
Cite this article
Liu, W., Li, Y., Learn, G. et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467, 420–425 (2010). https://doi.org/10.1038/nature09442
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09442
This article is cited by
-
Malaria-driven adaptation of MHC class I in wild bonobo populations
Nature Communications (2023)
-
Why Plasmodium vivax and Plasmodium falciparum are so different? A tale of two clades and their species diversities
Malaria Journal (2022)
-
A low-cost genomics workflow enables isolate screening and strain-level analyses within microbiomes
Genome Biology (2022)
-
Zoonotic origin of the human malaria parasite Plasmodium malariae from African apes
Nature Communications (2022)
-
Exposure of Primate Reservoir Hosts to Mosquito Vectors in Malaysian Borneo
EcoHealth (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.