Abstract
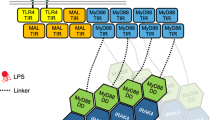
MyD88, IRAK4 and IRAK2 are critical signalling mediators of the TLR/IL1-R superfamily. Here we report the crystal structure of the MyD88–IRAK4–IRAK2 death domain (DD) complex, which surprisingly reveals a left-handed helical oligomer that consists of 6 MyD88, 4 IRAK4 and 4 IRAK2 DDs. Assembly of this helical signalling tower is hierarchical, in which MyD88 recruits IRAK4 and the MyD88–IRAK4 complex recruits the IRAK4 substrates IRAK2 or the related IRAK1. Formation of these Myddosome complexes brings the kinase domains of IRAKs into proximity for phosphorylation and activation. Composite binding sites are required for recruitment of the individual DDs in the complex, which are confirmed by mutagenesis and previously identified signalling mutations. Specificities in Myddosome formation are dictated by both molecular complementarity and correspondence of surface electrostatics. The MyD88–IRAK4–IRAK2 complex provides a template for Toll signalling in Drosophila and an elegant mechanism for versatile assembly and regulation of DD complexes in signal transduction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
O’Neill, L. A. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 226, 10–18 (2008)
Takeda, K., Kaisho, T. & Akira, S. Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 (2003)
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006)
Beutler, B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430, 257–263 (2004)
Verstak, B., Hertzog, P. & Mansell, A. Toll-like receptor signalling and the clinical benefits that lie within. Inflamm. Res. 56, 1–10 (2007)
Marx, J. Biomedicine. Puzzling out the pains in the gut. Science 315, 33–35 (2007)
O’Neill, L. A. Primer: Toll-like receptor signaling pathways–what do rheumatologists need to know? Nat. Clin. Pract. Rheumatol. 4, 319–327 (2008)
Marta, M., Meier, U. C. & Lobell, A. Regulation of autoimmune encephalomyelitis by toll-like receptors. Autoimmun. Rev. 8, 506–509 (2009)
Lande, R. et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 (2007)
Pisitkun, P. et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312, 1669–1672 (2006)
Horner, A. A. & Raz, E. Do microbes influence the pathogenesis of allergic diseases? Building the case for Toll-like receptor ligands. Curr. Opin. Immunol. 15, 614–619 (2003)
Chen, R. et al. Cancers take their Toll–the function and regulation of Toll-like receptors in cancer cells. Oncogene 27, 225–233 (2008)
Dasu, M. R., Devaraj, S., Park, S. & Jialal, I. Increased Toll-like receptor activation and TLR ligands in recently diagnosed type 2 diabetes subjects. Diabetes Care 33, 861–868 (2010)
Stewart, C. R. et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature Immunol. 11, 155–161 (2010)
den Dekker, W. K., Cheng, C., Pasterkamp, G. & Duckers, H. J. Toll like receptor 4 in atherosclerosis and plaque destabilization. Atherosclerosis 209, 314–320 (2010)
Romero-Sandoval, E. A., Horvath, R. J. & DeLeo, J. A. Neuroimmune interactions and pain: focus on glial-modulating targets. Curr. Opin. Investig. Drugs 9, 726–734 (2008)
O’Neill, L. A. & Bowie, A. G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Rev. Immunol. 7, 353–364 (2007)
Dinarello, C. A. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int. Rev. Immunol. 16, 457–499 (1998)
Kawai, T. et al. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11, 115–122 (1999)
Dunne, A. & O’Neill, L. A. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE 2003, re3 (2003)
Bowie, A. & O’Neill, L. A. The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J. Leukoc. Biol. 67, 508–514 (2000)
Beutler, B. et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24, 353–389 (2006)
Kobayashi, K. et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110, 191–202 (2002)
Suzuki, N. et al. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416, 750–756 (2002)
Kawagoe, T. et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nature Immunol. 9, 684–691 (2008)
Wan, Y. et al. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J. Biol. Chem. 284, 10367–10375 (2009)
Picard, C. et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299, 2076–2079 (2003)
von Bernuth, H. et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321, 691–696 (2008)
Lasker, M. V., Gajjar, M. M. & Nair, S. K. Cutting edge: Molecular structure of the IL-1R-associated kinase-4 death domain and its implications for TLR signaling. J. Immunol. 175, 4175–4179 (2005)
Motshwene, P. G. et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 284, 25404–25411 (2009)
Park, H. H. et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586 (2007)
Burns, K. et al. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197, 263–268 (2003)
Janssens, S. et al. MyD88S, a splice variant of MyD88, differentially modulates NF-κB- and AP-1-dependent gene expression. FEBS Lett. 548, 103–107 (2003)
Loiarro, M. et al. Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J. Biol. Chem. 284, 28093–28103 (2009)
Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 (1993)
Li, S., Strelow, A., Fontana, E. J. & Wesche, H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl Acad. Sci. USA 99, 5567–5572 (2002)
Szabo, G., Dolganiuc, A., Dai, Q. & Pruett, S. B. TLR4, ethanol, and lipid rafts: a new mechanism of ethanol action with implications for other receptor-mediated effects. J. Immunol. 178, 1243–1249 (2007)
Rao, N., Nguyen, S., Ngo, K. & Fung-Leung, W. P. A novel splice variant of interleukin-1 receptor (IL-1R)-associated kinase 1 plays a negative regulatory role in Toll/IL-1R-induced inflammatory signaling. Mol. Cell. Biol. 25, 6521–6532 (2005)
Hardy, M. P. & O’Neill, L. A. The murine Irak2 gene encodes four alternatively spliced isoforms, two of which are inhibitory. J. Biol. Chem. 279, 27699–27708 (2004)
Conze, D. B. et al. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol. Cell. Biol. 28, 3538–3547 (2008)
Cao, Z. et al. TRAF6 is a signal transducer for interleukin-1. Nature 383, 443–446 (1996)
Belvin, M. P. & Anderson, K. V. A conserved signaling pathway: the Drosophila Toll-Dorsal pathway. Annu. Rev. Cell Dev. Biol. 12, 393–416 (1996)
Sun, H., Bristow, B. N., Qu, G. & Wasserman, S. A. A heterotrimeric death domain complex in Toll signaling. Proc. Natl Acad. Sci. USA 99, 12871–12876 (2002)
Towb, P., Huaiyu, S. & Wasserman, S. A. Tube is an IRAK-4 homolog in a Toll pathway adapted for development and immunity. J. Innate Immun. 1, 309–321 (2009)
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995)
Moncrieffe, M. C., Grossmann, J. G. & Gay, N. J. Assembly of oligomeric death domain complexes during Toll receptor signaling. J. Biol. Chem. 283, 33447–33454 (2008)
Xiao, T., Towb, P., Wasserman, S. A. & Sprang, S. R. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell 99, 545–555 (1999)
Sun, H. et al. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. EMBO J. 23, 100–110 (2004)
Park, H. H. et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128, 533–546 (2007)
Egelman, E. H. Single-particle reconstruction from EM images of helical filaments. Curr. Opin. Struct. Biol. 17, 556–561 (2007)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Reynolds, C., Damerell, D. & Jones, S. ProtorP: a protein–protein interaction analysis server. Bioinformatics 25, 413–414 (2009)
Delano, W. L. The PyMol Molecular Graphics System 〈http://www.pymol.org/〉 (2002)
Acknowledgements
We thank K. Rajashankar, I. Kourinov and N. Sukumar for data collection and X. Ma for help with the manuscript. This work was supported by NIH (H.W.), the Cancer Research Institute (S.-C.L. and Y.-C.L.), and the American Heart Association (Y.-C.L.).
Author information
Authors and Affiliations
Contributions
H.W. initiated the project idea. S.-C.L. and Y.-C.L. designed and performed the experiments. S.-C.L. and H.W. interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4, Supplementary Figures 1-11 with legends and Supplementary Discussions 1-2. (PDF 10353 kb)
Rights and permissions
About this article
Cite this article
Lin, SC., Lo, YC. & Wu, H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 (2010). https://doi.org/10.1038/nature09121
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09121
This article is cited by
-
Role of innate immunological/inflammatory pathways in myelodysplastic syndromes and AML: a narrative review
Experimental Hematology & Oncology (2023)
-
PROTAC’ing oncoproteins: targeted protein degradation for cancer therapy
Molecular Cancer (2023)
-
Adipsin inhibits Irak2 mitochondrial translocation and improves fatty acid β-oxidation to alleviate diabetic cardiomyopathy
Military Medical Research (2023)
-
Emerging trends in IRAK-4 kinase research
Molecular Biology Reports (2023)
-
Targeting MyD88: Therapeutic mechanisms and potential applications of the specific inhibitor ST2825
Inflammation Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.