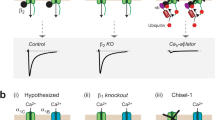
Abstract
Ca2+ channels and calmodulin (CaM) are two prominent signalling hubs1 that synergistically affect functions as diverse as cardiac excitability2, synaptic plasticity3 and gene transcription4. It is therefore fitting that these hubs are in some sense coordinated, as the opening of CaV1–2 Ca2+ channels are regulated by a single CaM constitutively complexed with channels5. The Ca2+-free form of CaM (apoCaM) is already pre-associated with the isoleucine–glutamine (IQ) domain on the channel carboxy terminus, and subsequent Ca2+ binding to this ‘resident’ CaM drives conformational changes that then trigger regulation of channel opening6. Another potential avenue for channel–CaM coordination could arise from the absence of Ca2+ regulation in channels lacking a pre-associated CaM6,7. Natural fluctuations in CaM concentrations might then influence the fraction of regulable channels and, thereby, the overall strength of Ca2+ feedback. However, the prevailing view has been that the ultrastrong affinity of channels for apoCaM ensures their saturation with CaM8, yielding a significant form of concentration independence between Ca2+ channels and CaM. Here we show that significant exceptions to this autonomy exist, by combining electrophysiology (to characterize channel regulation) with optical fluorescence resonance energy transfer (FRET) sensor determination of free-apoCaM concentration in live cells9. This approach translates quantitative CaM biochemistry from the traditional test-tube context into the realm of functioning holochannels within intact cells. From this perspective, we find that long splice forms of CaV1.3 and CaV1.4 channels include a distal carboxy tail10,11,12 that resembles an enzyme competitive inhibitor that retunes channel affinity for apoCaM such that natural CaM variations affect the strength of Ca2+ feedback modulation. Given the ubiquity of these channels13,14, the connection between ambient CaM levels and Ca2+ entry through channels is broadly significant for Ca2+ homeostasis. Strategies such as ours promise key advances for the in situ analysis of signalling molecules resistant to in vitro reconstitution, such as Ca2+ channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 April 2010
Nature 463, 968–972 (2010) In the Acknowledgements section of this Letter, the work was incorrectly listed as being funded in part by the US National Institute of Neurological Disorders and Stroke. This work was in fact funded by the US National Institute on Deafness and Other Communication Disorders.
References
Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabasi, A.-L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000)
Alseikhan, B. A., DeMaria, C. D., Colecraft, H. M. & Yue, D. T. Engineered calmodulins reveal the unexpected eminence of Ca2+ channel inactivation in controlling heart excitation. Proc. Natl Acad. Sci. USA 99, 17185–17190 (2002)
Xu, J. & Wu, L. G. The decrease in the presynaptic calcium current is a major cause of short-term depression at a calyx-type synapse. Neuron 46, 633–645 (2005)
Krey, J. F. & Dolmetsch, R. E. Molecular mechanisms of autism: a possible role for Ca2+ signaling. Curr. Opin. Neurobiol. 17, 112–119 (2007)
Yang, P. S., Mori, M. X., Antony, E. A., Tadross, M. R. & Yue, D. T. A single calmodulin imparts distinct N- and C-lobe regulatory processes to individual CaV1.3 channels. Biophys. J. Suppl. abstr. 1669-Plat (2007)
Erickson, M. G., Liang, H., Mori, M. X. & Yue, D. T. FRET two-hybrid mapping reveals function and location of L-type Ca2+ channel CaM preassociation. Neuron 39, 97–107 (2003)
Liang, H. et al. Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron 39, 951–960 (2003)
Yang, P. S. et al. Switching of Ca2+-dependent inactivation of CaV1.3 channels by calcium binding proteins of auditory hair cells. J. Neurosci. 26, 10677–10689 (2006)
Black, D. J., Leonard, J. & Persechini, A. Biphasic Ca2+-dependent switching in a calmodulin-IQ domain complex. Biochemistry 45, 6987–6995 (2006)
Singh, A. et al. Modulation of voltage- and Ca2+-dependent gating of CaV1.3 L-type calcium channels by alternative splicing of a C-terminal regulatory domain. J. Biol. Chem. 283, 20733–20744 (2008)
Singh, A. et al. C-terminal modulator controls Ca2+-dependent gating of CaV1.4 L-type Ca2+ channels. Nature Neurosci. 9, 1108–1116 (2006)
Wahl-Schott, C. et al. Switching off calcium-dependent inactivation in L-type calcium channels by an autoinhibitory domain. Proc. Natl Acad. Sci. USA 103, 15657–15662 (2006)
Platzer, J. et al. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell 102, 89–97 (2000)
Namkung, Y. et al. Requirement for the L-type Ca2+ channel α1D subunit in postnatal pancreatic β cell generation. J. Clin. Invest. 108, 1015–1022 (2001)
Strom, T. M. et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nature Genet. 19, 260–263 (1998)
Doering, C. J., Hamid, J., Simms, B., McRory, J. E. & Zamponi, G. W. CaV1.4 encodes a calcium channel with low open probability and unitary conductance. Biophys. J. 89, 3042–3048 (2005)
Dick, I. E. et al. A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature 451, 830–834 (2008)
Peterson, B. Z. et al. Critical determinants of Ca2+-dependent inactivation within an EF-hand motif of L-type Ca2+ channels. Biophys. J. 78, 1906–1920 (2000)
Griessmeier, K. et al. Calmodulin is a functional regulator of CaV1.4 L-type Ca2+ channels. J. Biol. Chem. 284, 29809–29816 (2009)
Cantor, C. R. & Schimmel, P. R. Biophysical Chemistry: Part III: The Behavior of Biological Macromolecules 11th edn, 887–978 (Macmillan, 1980)
Kim, J., Ghosh, S., Nunziato, D. A. & Pitt, G. S. Identification of the components controlling inactivation of voltage-gated Ca2+ channels. Neuron 41, 745–754 (2004)
Xiong, L., Kleerekoper, Q. K., He, R., Putkey, J. A. & Hamilton, S. L. Sites on calmodulin that interact with the C-terminal tail of CaV1.2 channel. J. Biol. Chem. 280, 7070–7079 (2005)
Black, D. J., Tran, Q. K. & Persechini, A. Monitoring the total available calmodulin concentration in intact cells over the physiological range in free Ca2+ . Cell Calcium 35, 415–425 (2004)
Slemmon, J. R., Feng, B. & Erhardt, J. A. Small proteins that modulate calmodulin-dependent signal transduction: effects of PEP-19, neuromodulin, and neurogranin on enzyme activation and cellular homeostasis. Mol. Neurobiol. 22, 99–113 (2000)
Chambers, J. S., Thomas, D., Saland, L., Neve, R. L. & Perrone-Bizzozero, N. I. Growth-associated protein 43 (GAP-43) and synaptophysin alterations in the dentate gyrus of patients with schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 29, 283–290 (2005)
Bezprozvanny, I. Calcium signaling and neurodegenerative diseases. Trends Mol. Med. 15, 89–100 (2009)
Masliah, E. et al. Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science 287, 1265–1269 (2000)
Lee, D., Lee, S. Y., Lee, E. N., Chang, C. S. & Paik, S. R. α-Synuclein exhibits competitive interaction between calmodulin and synthetic membranes. J. Neurochem. 82, 1007–1017 (2002)
Chan, C. S. et al. ‘Rejuvenation’ protects neurons in mouse models of Parkinson’s disease. Nature 447, 1081–1086 (2007)
Ikeda, S. et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol. Cell. Biol. 29, 2193–2204 (2009)
Acknowledgements
We thank M. Tadross, I. Dick and members of the Calcium Signals Laboratory for comments; M. Tadross for data-acquisition software; D. J. Black and A. Persechini for BSCaMIQ and neuromodulin complementary DNA; J. McRory and T. Snutch for human α1F cDNA; J. Streissnig for human α1D cDNA; and V. Wu for earlier foundational experiments. This work is supported by grants from the US National Institute of Mental Health, the US National Heart, Lung, and Blood Institute and the US National Institute of Neurological Disorders and Stroke.
Author Contributions X.L. devised and refined experimental design, carried out all phases of the experiments and performed extensive data analysis. P.S.Y. consulted on initial molecular biology approaches, constructed certain channels with ICDI point mutations and contributed importantly to CaV1.4 expression strategies and electrophysiological characterization. W.Y. conducted FRET experiments, undertook molecular biology and extensively managed technical aspects of the project. D.T.Y. conceived, refined and oversaw the experiments, performed FRET experiments, analysed data and wrote the manuscript. All authors commented on and edited the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Information and Data including Supplementary Figures S1.1- S1.5, S2.2-S2.3, S3.2, S3.4-S3.6 & S4.2 with Legends and Supplementary References. (PDF 3710 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Yang, P., Yang, W. et al. Enzyme-inhibitor-like tuning of Ca2+ channel connectivity with calmodulin. Nature 463, 968–972 (2010). https://doi.org/10.1038/nature08766
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08766
This article is cited by
-
Cytosolic peptides encoding CaV1 C-termini downregulate the calcium channel activity-neuritogenesis coupling
Communications Biology (2022)
-
Cav1.4 dysfunction and congenital stationary night blindness type 2
Pflügers Archiv - European Journal of Physiology (2021)
-
Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP
Nature Communications (2018)
-
Calmodulin interacts with Rab3D and modulates osteoclastic bone resorption
Scientific Reports (2016)
-
Splice variants of the CaV1.3 L-type calcium channel regulate dendritic spine morphology
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.