Abstract
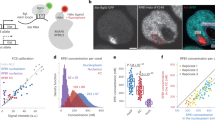
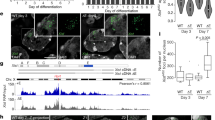
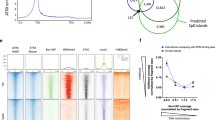
Nuclear compartmentalization seems to have an important role in regulating metazoan genes1,2. Although studies on immunoglobulin and other loci have shown a correlation between positioning at the nuclear lamina and gene repression, the functional consequences of this compartmentalization remain untested2,3. We devised an approach for inducible tethering of genes to the inner nuclear membrane (INM), and tested the consequences of such repositioning on gene activity in mouse fibroblasts. Here, using three-dimensional DNA-immunoFISH, we demonstrate repositioning of chromosomal regions to the nuclear lamina that is dependent on breakdown and reformation of the nuclear envelope during mitosis. Moreover, tethering leads to the accumulation of lamin and INM proteins, but not to association with pericentromeric heterochromatin or nuclear pore complexes. Recruitment of genes to the INM can result in their transcriptional repression. Finally, we use targeted adenine methylation (DamID) to show that, as is the case for our model system, inactive immunoglobulin loci at the nuclear periphery are contacted by INM and lamina proteins. We propose that these molecular interactions may be used to compartmentalize and to limit the accessibility of immunoglobulin loci to transcription and recombination factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The microarray data can be found at the Gene Expression Omnibus at NCBI (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE10176.
References
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G. & Cremer, T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8, 104–115 (2007)
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007)
Kosak, S. T. et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162 (2002)
Hewitt, S. L., High, F. A., Reiner, S. L., Fisher, A. G. & Merkenschlager, M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur. J. Immunol. 34, 3604–3613 (2004)
Zink, D. et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166, 815–825 (2004)
Chuang, C.-H. et al. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825–831 (2006)
Williams, R. R. et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119, 132–140 (2006)
Andrulis, E. D., Neiman, A. M., Zappulla, D. C. & Sternglanz, R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394, 592–595 (1998)
Taddei, A. et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778 (2006)
Schmid, M. et al. Nup-PI: The nucleopore–promoter interaction of genes in yeast. Mol. Cell 21, 379–391 (2006)
Shaklai, S., Amariglio, N., Rechavi, G. & Simon, A. J. Gene silencing at the nuclear periphery. FEBS J. 274, 1383–1392 (2007)
Somech, R. et al. The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 118, 4017–4025 (2005)
Holaska, J. M., Lee, K. K., Kowalski, A. K. & Wilson, K. L. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 278, 6969–6975 (2003)
Polioudaki, H. et al. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2, 920–925 (2001)
Belmont, A. S., Li, G., Sudlow, G. & Robinett, C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell Biol. 58, 203–222 (1999)
Tsuchiya, Y., Hase, A., Ogawa, M., Yorifuji, H. & Arahata, K. Distinct regions specify the nuclear membrane targeting of emerin, the responsible protein for Emery–Dreifuss muscular dystrophy. Eur. J. Biochem. 259, 859–865 (1999)
Yang, Q., Riblet, R. & Schildkraut, C. L. Sites that direct nuclear compartmentalization are near the 5′ end of the mouse immunoglobulin heavy-chain locus. Mol. Cell. Biol. 25, 6021–6030 (2005)
Vogel, M. J., Peric-Hupkes, D. & van Steensel, B. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nature Protocols 2, 1467–1478 (2007)
Bengtsson, L. & Wilson, K. L. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr. Opin. Cell Biol. 16, 73–79 (2004)
Pickersgill, H. et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature Genet. 38, 1005–1014 (2006)
Bertolino, E. et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nature Immunol. 6, 836–843 (2005)
Johnson, K., Angelin-Duclos, C., Park, S. & Calame, K. L. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V–DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 23, 2438–2450 (2003)
Pawlitzky, I. et al. Identification of a candidate regulatory element within the 5′ flanking region of the mouse Igh locus defined by pro-B cell-specific hypersensitivity associated with binding of PU.1, Pax5, and E2A. J. Immunol. 176, 6839–6851 (2006)
Ragoczy, T., Bender, M. A., Telling, A., Byron, R. & Groudine, M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20, 1447–1457 (2006)
Mendjan, S. et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21, 811–823 (2006)
Kim, S. H. et al. Spatial genome organization during T-cell differentiation. Cytogenet. Genome Res. 105, 292–301 (2004)
Solovei, I. et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 276, 10–23 (2002)
Jolly, C., Mongelard, F., Robert-Nicoud, M. & Vourc’h, V. Optimization of nuclear transcript detection by FISH and combination with fluorescence immunocytochemical detection of transcription factors. J. Histochem. Cytochem. 45, 1585–1592 (1997)
Acknowledgements
We thank A. Belmont for the lacO and GFP-LacI plasmids and K. Van Steensel for DamID plasmids. We are grateful to members of the laboratory for critical input and support by the Howard Hughes Medical Institute. J.M.Z. is supported by an NIH training grant.
Author Contributions K.L.R. designed and performed most of the experiments. J.M.Z. carried out the DamID experiments with K.L.R.. E.B. contributed the microarray analysis. K.L.R. and H.S. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Figures S1-S7 with Legends, Supplementary Methods and additional references. (PDF 1576 kb)
Rights and permissions
About this article
Cite this article
Reddy, K., Zullo, J., Bertolino, E. et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 (2008). https://doi.org/10.1038/nature06727
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06727
This article is cited by
-
From the membrane to the nucleus: mechanical signals and transcription regulation
Biophysical Reviews (2023)
-
The regional sequestration of heterochromatin structural proteins is critical to form and maintain silent chromatin
Epigenetics & Chromatin (2022)
-
Histone N-terminal acetyltransferase NAA40 links one-carbon metabolism to chemoresistance
Oncogene (2022)
-
Lamin A/C-dependent chromatin architecture safeguards naïve pluripotency to prevent aberrant cardiovascular cell fate and function
Nature Communications (2022)
-
Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.