Abstract
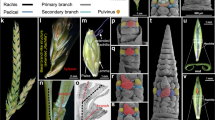
The external appearance of flowering plants is determined to a large extent by the forms of flower-bearing branch systems, known as inflorescences, and their position in the overall structure of the plant. Branches and branching patterns are produced by tissues called shoot apical meristems. Thus, inflorescence architecture reflects meristem number, arrangement and activity, and the duration of meristem activity correlates with branch length. The inflorescences of maize, unlike those of related grasses such as rice and sorghum, predominantly lack long branches, giving rise to the tassel and familiar corncob. Here we report the isolation of the maize ramosa1 gene and show that it controls inflorescence architecture. Through its expression in a boundary domain near the nascent meristem base, ramosa1 imposes short branch identity as branch meristems are initiated. A second gene, ramosa2, acts through ramosa1 by regulating ramosa1 gene expression levels. ramosa1 encodes a transcription factor that appears to be absent in rice, is heterochronically expressed in sorghum, and may have played an important role in maize domestication and grass evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Weberling, F. Morphology of Flowers and Inflorescences (Cambridge Univ. Press, Cambridge, 1989)
Sussex, I. M. & Kerk, N. M. The evolution of plant architecture. Curr. Opin. Plant Biol. 4, 33–37 (2001)
Clifford, H. in Grass Systematics and Evolution (eds Soderstrom, T., Hilu, K., Campbell, C. & Barkworth, M.) 21–30 (Smithsonian Institution Press, Washington DC, 1987)
Doust, A. N. & Kellogg, E. A. Inflorescence diversification in the panicoid “bristle grass” clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology. Am. J. Bot. 89, 1203–1222 (2002)
McSteen, P., Laudencia-Chingcuanco, D. & Colasanti, J. A floret by any other name: control of meristem identity in maize. Trends Plant Sci. 5, 61–66 (2000)
Veit, B., Schmidt, R. J., Hake, S. & Yanofsky, M. F. Maize floral development: new genes and old mutants. Plant Cell 5, 1205–1215 (1993)
Gernert, W. A new subspecies of Zea mays L. Am. Nat. 46, 616–622 (1912)
Chuck, G., Muszynski, M., Kellogg, E., Hake, S. & Schmidt, R. J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298, 1238–1241 (2002)
Postlethwait, S. N. & Nelson, O. E. Characterization of development in maize through the use of mutants. I. The polytypic (Pt) and ramosa-1 (ra1) mutants. Am. J. Bot. 51, 238–243 (1964)
Emerson, R., Beadle, G. & Fraser, A. A summary of linkage studies in maize. Cornell Univ. Agric. Experiment Station Memoir 180, 3–83 (1935)
Cheng, P. C., Greyson, R. I. & Walden, D. B. Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 70, 450–462 (1983)
McClintock, B. Induction of instability at selected loci in maize. Genetics 38, 579–599 (1953)
Dawe, R. K. & Freeling, M. Clonal analysis of the cell lineages in the male flower of maize. Dev. Biol. 142, 233–245 (1990)
Takatsuji, H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078 (1999)
Sakai, H., Medrano, L. J. & Meyerowitz, E. M. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203 (1995)
Nickerson, N. H. & Dale, E. E. Tassel modifications in Zea mays. Ann. Mo. Bot. Gard. 42, 195–211 (1955)
Walsh, J. & Freeling, M. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot. Plant J. 19, 489–495 (1999)
Kellogg, E. in Grasses: Systematics and Evolution (eds Jacobs, S. & Everett, J.) (CSIRO, Melbourne, 2000)
Kellogg, E. A. Plant evolution: the dominance of maize. Curr. Biol. 7, R411–R413 (1997)
Whitt, S. R., Wilson, L. M., Tenaillon, M. I., Gaut, B. S. & Buckler, E. S. Genetic diversity and selection in the maize starch pathway. Proc. Natl Acad. Sci. USA 99, 12959–12962 (2002)
Hudson, R., Kreitman, M. & Aguade, M. A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 (1987)
Tenaillon, M. I. et al. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl Acad. Sci. USA 98, 9161–9166 (2001)
Remington, D. L. et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl Acad. Sci. USA 98, 11479–11484 (2001)
Wilson, L. M. et al. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16, 2719–2733 (2004)
Troll, W. Die Infloreszenzen: Typologie und Stellung im Aufbau des Vegetationskörpers (Fischer, Stuttgart, 1964)
Poethig, R. Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930 (1990)
Gallavotti, A. et al. The role of barren stalk1 in the architecture of maize. Nature 432, 630–635 (2004)
Doebley, J., Stec, A. & Hubbard, L. The evolution of apical dominance in maize. Nature 386, 485–488 (1997)
Chuck, G., Meeley, R. & Hake, S. The control of maize spikelet meristem identity by the APETALA-2-like gene indeterminate spikelet1. Genes Dev. 12, 1145–1154 (1998)
Taguchi-Shiobara, F., Yuan, Z., Hake, S. & Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15, 2755–2766 (2001)
Bommert, P. et al. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132, 1235–1245 (2005)
Vollbrecht, E., Reiser, L. & Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172 (2000)
Collins, G. Hybrids of Zea tunicata and Zea ramosa. Proc. Natl Acad. Sci. USA 3, 345–349 (1917)
Doebley, J. & Stec, A. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134, 559–570 (1993)
Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. The limits of selection during maize domestication. Nature 398, 236–239 (1999)
Takeda, T. et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520 (2003)
Doebley, J. The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59 (2004)
Kellogg, E. A. Evolution of developmental traits. Curr. Opin. Plant Biol. 7, 92–98 (2004)
Martienssen, R. The origin of maize branches out. Nature 386, 443–444 (1997)
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2001)
Jackson, D., Veit, B. & Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413 (1994)
Rozas, J. & Rozas, R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175 (1999)
Acknowledgements
We thank T. Mulligan for plant care, Z. Lippman and C. Kopec for help with in situ hybridization and SEM, R. J. Schmidt for producing and sharing the ra1-RS allele, D. Jackson for discussions and V. Irish for commenting on the manuscript. E.V. was a DOE-Energy Biosciences postdoctoral fellow of the Life Sciences Research Foundation. L.G. was supported by the Cold Spring Harbor Undergraduate Research Program. Grant support was provided by the Agricultural Research Service of the USDA (to E.S.B.), the National Research Initiative of the USDA CSREES (to R.M.), and by the NSF Plant Genome Research Program (to E.S.B. and R.M.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
DNA sequences reported here have been deposited in GenBank under accession numbers AY957396–AY957399 and DQ013174–DQ013203. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table S1
This Microsoft Word format file contains a table with statistics of nucleotide diversity and HKA tests. (DOC 30 kb)
Supplementary Methods
This Microsoft Word format file contains text that summarizes genetic methods for cosegregation analysis of mutable, Spm-induced alleles of ramosa1. (DOC 24 kb)
Rights and permissions
About this article
Cite this article
Vollbrecht, E., Springer, P., Goh, L. et al. Architecture of floral branch systems in maize and related grasses. Nature 436, 1119–1126 (2005). https://doi.org/10.1038/nature03892
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature03892
This article is cited by
-
Transposable element-initiated enhancer-like elements generate the subgenome-biased spike specificity of polyploid wheat
Nature Communications (2023)
-
QTG-Miner aids rapid dissection of the genetic base of tassel branch number in maize
Nature Communications (2023)
-
Deploying QTL-seq rapid identification and separation of the major QTLs of tassel branch number for fine-mapping in advanced maize populations
Molecular Breeding (2023)
-
Genetic architecture of ear traits based on association mapping and co-expression networks in maize inbred lines and hybrids
Molecular Breeding (2023)
-
Transcriptional Signatures in Contrasting Cultivars of Barley During Early Reproductive Development of Meristem
Journal of Plant Growth Regulation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.