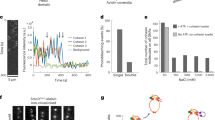
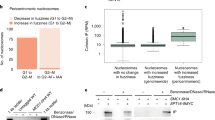
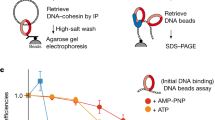
Abstract
Sister chromatids, the products of eukaryotic DNA replication, are held together by the chromosomal cohesin complex after their synthesis. This allows the spindle in mitosis to recognize pairs of replication products for segregation into opposite directions1,2,3,4,5,6. Cohesin forms large protein rings that may bind DNA strands by encircling them7, but the characterization of cohesin binding to chromosomes in vivo has remained vague. We have performed high resolution analysis of cohesin association along budding yeast chromosomes III–VI. Cohesin localizes almost exclusively between genes that are transcribed in converging directions. We find that active transcription positions cohesin at these sites, not the underlying DNA sequence. Cohesin is initially loaded onto chromosomes at separate places, marked by the Scc2/Scc4 cohesin loading complex8, from where it appears to slide to its more permanent locations. But even after sister chromatid cohesion is established, changes in transcription lead to repositioning of cohesin. Thus the sites of cohesin binding and therefore probably sister chromatid cohesion, a key architectural feature of mitotic chromosomes, display surprising flexibility. Cohesin localization to places of convergent transcription is conserved in fission yeast, suggesting that it is a common feature of eukaryotic chromosomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Michaelis, C., Ciosk, R. & Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45 (1997)
Guacci, V., Koshland, D. & Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell 91, 47–57 (1997)
Losada, A., Hirano, M. & Hirano, T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12, 1986–1997 (1998)
Tomonaga, T. et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14, 2757–2770 (2000)
Tanaka, T., Fuchs, J., Loidl, J. & Nasmyth, K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol. 2, 492–499 (2000)
Uhlmann, F. Chromosome cohesion and separation: From men and molecules. Curr. Biol. 13, R104–R114 (2003)
Haering, C. H., Löwe, J., Hochwagen, A. & Nasmyth, K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 (2002)
Ciosk, R. et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 1–20 (2000)
Blat, Y. & Kleckner, N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98, 249–259 (1999)
Tanaka, T., Cosma, M. P., Wirth, K. & Nasmyth, K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98, 847–858 (1999)
Megee, P. C. & Koshland, D. A functional assay for centromere-associated sister chromatid cohesion. Science 285, 254–257 (1999)
Laloraya, S., Guacci, V. & Koshland, D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151, 1047–1056 (2000)
Nonaka, N. et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell Biol. 4, 89–93 (2001)
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001)
Hakimi, M.-A. et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418, 994–997 (2002)
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003)
Filipski, J. & Mucha, M. Structure, function and DNA composition of Saccharomyces cerevisiae chromatin loops. Gene 300, 63–68 (2002)
Chu, S. et al. The transcriptional program of sporulation in budding yeast. Science 282, 699–705 (1998)
Gasch, A. P. et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000)
Tóth, A. et al. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320–333 (1999)
Arumugam, P. et al. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 13, 1941–1953 (2003)
Sjögren, C. & Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991–995 (2001)
Jacq, C. et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature 387(suppl), 75–78 (1997)
Donze, D., Adams, C. R., Rine, J. & Kamakaka, R. T. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13, 698–708 (1999)
Cohen, B. A., Mitra, R. D., Hughes, J. D. & Church, G. M. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nature Genet. 26, 183–186 (2000)
Weitzer, S., Lehane, C. & Uhlmann, F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 13, 1930–1940 (2003)
Toyoda, Y. et al. Requirement of chromatid cohesion proteins Rad21/Scc1 and Mis4/Scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12, 347–358 (2002)
Waizenegger, I. C., Hauf, S., Meinke, A. & Peters, J.-M. Two distinct pathways remove mammalian cohesin complexes from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 (2000)
Reid, R. J. D., Sunjevaric, I., Kedacche, M. & Rothstein, R. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19, 319–328 (2002)
Ohta, K. et al. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc. Natl Acad. Sci. USA 95, 645–651 (1998)
Acknowledgements
We are indebted to E. Schwob for initiating this collaboration. We also thank A. Nakada and T. Chaplin for technical support, R. Rothstein for reagents, and J. Cau, J. Sgouros, J. Svejstrup and members of our laboratories for discussions and comments on the manuscript. A.L. was supported by an EU Marie Curie individual fellowship and a Journal of Cell Science travelling fellowship; K.S. acknowledges support through a MEXT grants-in-aid for priority areas; F.U. was supported by the EMBO Young Investigator programme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure 1
Binding of cohesin subunits to S. cerevisiae chromosome VI. (PDF 170 kb)
Supplementary Figure 2
Binding of the cohesin subunit Scc1 to S. cerevisiae chromosomes III-V at 23ºC. (PDF 821 kb)
Supplementary Figure 3
Changes in the binding of Scc1 to S. cerevisiae chromosome VI in response to transcription. (PDF 372 kb)
Supplementary Figure 4
Binding of the cohesin subunit Scc1 to S. cerevisiae chromosomes III-V after heat-shock. (PDF 769 kb)
Supplementary Figure 5
Binding of Scc2, Scc4 and transcriptional activity along S. cerevisiae chromosome VI. (PDF 150 kb)
Supplementary Figure 6
Loading of cohesin at Scc2 binding sites on S. cerevisiae chromosome VI. (PDF 146 kb)
Supplementary Note 1
Relationship of Scc2 binding and transcriptional activity along S. cerevisiae chromosome VI. (PDF 98 kb)
Supplementary Note 2
Cohesin association sites on S.pombe chromosomes 2-3. (PDF 384 kb)
Supplementary Note 3
Gene orientation analysis on S. cerevisiae chromosomes III-VI. (PDF 60 kb)
Supplementary Table 1
Cohesin association sites on S. cerevisiae chromosomes III-VI. (PDF 37 kb)
Supplementary Table 2
Strain list. (PDF 52 kb)
Rights and permissions
About this article
Cite this article
Lengronne, A., Katou, Y., Mori, S. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 (2004). https://doi.org/10.1038/nature02742
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature02742
This article is cited by
-
Oncogenic c-Myc induces replication stress by increasing cohesins chromatin occupancy in a CTCF-dependent manner
Nature Communications (2024)
-
RAD21 is the core subunit of the cohesin complex involved in directing genome organization
Genome Biology (2023)
-
Different NIPBL requirements of cohesin-STAG1 and cohesin-STAG2
Nature Communications (2023)
-
DNA double-strand break end synapsis by DNA loop extrusion
Nature Communications (2023)
-
The multifaceted roles of cohesin in cancer
Journal of Experimental & Clinical Cancer Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.