Abstract
Recent data suggest that detection of epidermal growth factor receptor protein by immunohistochemistry (IHC) does not predict response to the antiepidermal growth factor receptor drug, cetuximab, in patients with colorectal carcinoma. In searching for foundation for further investigation to optimize patient selection for cetuximab therapy, this study sought to exploit the tissue microarray and chromogenic in situ hybridization techniques to evaluate the status of epidermal growth factor receptor gene amplification in colorectal cancer and its relationship with protein expression by IHC. The study included 158 primary or metastatic colorectal adenocarcinomas. Immunohistochemical results were scored as 0–3+ based on the intensity of membrane staining. The in situ hybridization signals were counted in 30 nuclei per tissue core. Overall, the rate of tissue loss was 7%, yielding 147 analyzable cases: 123 primary, 24 metastatic. Positive immunohistochemical staining of any intensity was detected in 85% (105/123) of primary and 79% (19/24) of metastatic tumors, whereas gene amplification (>5 gene copies/nucleus) was only seen in 12% (15/123) of primary and 8% (2/24) of metastatic tumors. Only 2/15 primary and 1/2 metastatic tumors that showed gene amplification were amplified at a high level (>10 gene copies/nucleus). Although a positive correlation was detected between the intensity of protein expression and the likelihood of gene amplification in both the primary (P=0.01) and the metastatic (P=0.05) tumors, IHC had a low specificity (17% in primary, 23% in metastatic) in predicting gene amplification. Conversely, all tumors that did not express the protein by IHC lacked gene amplification. Thus, this study shows that only a small fraction of epidermal growth factor receptor- positive colorectal carcinomas detected by IHC are associated with gene amplification. Additional studies are needed to determine whether epidermal growth factor receptor gene amplification bears any informative value in predicting response to cetuximab-based therapy.
Similar content being viewed by others
Main
The epidermal growth factor receptor (EGFR) is a member of the HER tyrosine kinase growth factor receptor family, and is involved in signaling pathways affecting cell growth.1, 2, 3, 4 Recent studies have shown that EGFR expression is present in approximately 60–80% of colorectal carcinomas5, 6 and the receptor has emerged as a rational target for anticancer therapy in these tumors.2, 7 Cetuximab is a human–murine chimeric monoclonal antibody that specifically blocks the EGFR. Several clinical trials have demonstrated activity of cetuximab in patients with advanced colorectal carcinoma3, 4 and the drug is currently licensed in the US and Switzerland for use in such patients.
Clinical trials on the use of cetuximab in patients with colorectal carcinoma have been performed exclusively on EGFR-positive tumors as detected by immunohistochemistry (IHC). This decision was based on the finding from preclinical studies that EGFR expression is needed for cetuximab activity. Consequently, immunohistochemical positivity for EGFR has been used as a criterion for patient selection. It has been noted, however, that no apparent relationship exists between the efficacy of cetuximab and the extent of EGFR staining in the tumor.8, 9, 10, 11 Furthermore, response to cetuximab has recently been shown in patients with EGFR-negative colorectal carcinomas.12, 13 Thus, the value of immunohistochemical detection of EGFR in selecting patients for treatment with cetuximab has failed to be borne in our clinical trails. An alternative methodology that offers higher sensitivity and specificity in predicting treatment response would be highly desirable.
Chromogenic in situ hybridization is a technique that can be used to evaluate gene amplification or deletion, chromosome aneuploidy, or chromosome translocation on tissue sections.14, 15, 16 Studies on its use in archival breast cancer specimens have shown it to be a practical, cost-effective and valid alternative to fluorescent in situ hybridization in detecting gene amplification.14, 17 Instead of using a fluorescent dye, chromogenic in situ hybridization uses a peroxidase reaction to detect the gene probe. The hybridization result can be visualized under a routine light microscope. Like fluorescent in situ hybridization, chromogenic in situ hybridization is performed on routine paraffin tissue sections; the gene signals are analyzed in the context of morphology and can be easily interpreted by surgical pathologists. Gene amplification can also be reliably identified in tissue sections where carcinoma cells are admixed with abundant non-neoplastic components. The use of chromogenic in situ hybridization in the evaluation of EGFR has been explored in archival gliomas,18 but not in colorectal tumors. In gliomas, all tumors with EGFR gene amplification showed high protein expression.
Tissue microarray technique is also a relatively new technique. It allows rapid, high throughput expression profiling by IHC in archival tissues19, 20, 21 and its utility has been validated by several studies using both epithelial and mesenchymal tumor samples.22, 23, 24 More recently, tissue microarray has also been shown to be an effective method for the assessment of HER-2 gene amplification with fluorescent in situ hybridization or chromogenic in situ hybridization in breast tumors.25, 26
In searching for foundations for further investigation to optimize patient selection for cetuximab therapy, we sought to exploit both the chromogenic in situ hybridization and tissue microarray techniques, and to analyze the frequency of EGFR gene amplification and its relationship with immunohistochemical protein expression in colorectal carcinomas by performing a comparative immunohistochemical and chromogenic in situ hybridization study on tissue microarray sections. We anticipated that results from such a study would help determine the feasibility of further clinical trials that use chromogenic in situ hybridization or similar techniques to predict treatment response to anti-EGFR therapy.
Materials and methods
Tumor Samples and Construction of Tissue Microarray
A total of 158 primary (130) and metastatic (28) colorectal, adenocarcinomas derived from 158 patients were randomly selected for this study. Two tissue microarrays were constructed using 0.6 mm tissue cores as previously described.22 A hematoxylin and eosin (H&E)-stained section was evaluated for the presence of carcinoma and the area to be used for creation of the tissue microarray was marked on the slide and the donor block. Three cores of different areas of the tumor from a single formalin-fixed, paraffin-embedded tissue block were sampled.
Immunohistochemistry
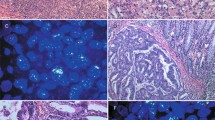
Immunohistochemical staining for EGFR was performed on the tissue microarray slides using the EGFR pharmDx kit (DakoCytomation, Carpentino, CA, USA) according to the manufacturer's instructions. The stains were scored as 0 when there was no specific membrane staining within the tumor, and positive when there was any staining of tumor cell membrane above background level. The positive cases were further classified into 1+, 2+ and 3+ based on the staining intensity. The highest staining intensity of all tissue cores from the same tumor was used as the final immunohistochemical result for that tumor. Examples of negative and positive stains are illustrated in Figure 1a–d.
Chromogenic In Situ Hybridization
Chromogenic in situ hybridization for the EGFR gene was performed according to the manufacturer's (Zymed Laboratories Inc., South San Francisco, CA, USA) instructions. Briefly, the tissue microarray sections were incubated at 55°C overnight. The slides were deparaffinized in xylene and graded ethanols. Heat pretreatment was carried out in the pretreatment buffer (Zymed Laboratories Inc.) at 98–100°C for 15 min. The tissue was digested with pepsin for 10 min at room temperature. After application of Zymed SpotLight® digoxigenin labeled EGFR probe (Zymed Laboratories Inc.), the slides were coverslipped and edges sealed with rubber cement. The slides were heated at 95°C for 5 min followed by overnight incubation at 37°C using a moisturized chamber. A posthybridization wash was performed the next day and followed by immunodetection using the CISH™ polymer detection kit (Zymed Laboratories Inc.). Signal enumeration was performed under a standard light microscope using a × 40 objective. The total gene copy numbers in 30 nuclei were counted for each tissue core, and the average gene copies per nucleus were used as results for that tissue core. The highest chromogenic in situ hybridization score among all cores was used as the final result for that tumor. The results were interpreted as follows: <5 gene copies per nucleus, no amplification; 5–10 gene copies per nucleus, low-level amplification; and >10 gene copies per nucleus, high-level amplification. Staining examples are illustrated in Figure 2a–d.
Statistical Analyses
Correlation between the immunohistochemical staining intensity and the level of gene amplification was tested using the Jonckheere–Terpstra test,27 and a P-value ≤0.05 was considered significant.
Results
Tumors with at least two tissue cores available for scoring were included in the final analyses. Eleven of 158 (7%) tumors were lost during processing, including seven primary and four metastatic tumors. The reasons for failure were complete loss of tissue cores, less than 30 tumor cells available for scoring in a tissue core, and absence of EGFR signal. The absence of signal probably resulted from under- or overdigestion, since tissue digestion for a particular case cannot be controlled on a tissue microarray.
Of the147 analyzable tumors, 123 were primary and 24 were metastatic (six liver metastases, and 18 lung metastases) tumors. Overall, EGFR immunohistochemical staining of any intensity was observed in 105/123 (85%) primary tumors and 19/24 (79%) metastatic tumors (Table 1). Of the 105 positive primary carcinomas, the staining intensity was 1+ in 40/105 (38%) cases, 2+ in 45/105 (43%), and 3+ in 20/105 (19%). Of the 19 positive metastatic cases, the staining intensity was 1+ in 10 (53%) cases, 2+ in eight (42%), and 3+ in one (5%). In contrast, gene amplification by chromogenic in situ hybridization was present in only 15 of 123 (12%) primary tumors and two of 24 (8%) metastatic tumors. Furthermore, only two primary tumors and one metastatic tumor showed high-level EGFR gene amplification. All primary and metastatic tumors that had gene amplification showed positive immunohistochemical staining, and there was a positive correlation between the intensity of immunohistochemical positivity and gene amplification in both the primary (P=0.01) and metastatic (P=0.05) tumors. However, the specificity of IHC in predicting gene amplification was extremely low, as 90 of 108 (83%) nonamplified primary tumors, and 17 of 22 (77%) nonamplified metastatic tumors showed positive immunohistochemical staining.
Discussion
The reasons why immunohistochemical detection of EGFR is a poor indicator of response to cetuximab may include a variety of biological and technical factors. The most critical concern is that the EGFR detected by IHC is not the form acted upon by cetuximab.13 From a biological point of view, the EGFR signaling pathway is complex,28 and it is possible that the level of expression of the receptor's ligands, the level of tyrosine phosphorylation of the receptor, or the expression of other downstream molecules, are critically involved in the action of cetuximab and therefore more predictive of treatment response than the total level of the receptor per se as detected by IHC. From a technical point of view, tissue processing and handling,29, 30 the prolonged storage time of tissue samples30 may allow certain catalytic degradation of cell surface receptors, resulting in altered protein expression. Furthermore, the clone of antibody used for IHC might also have different specificities for certain epitopes, and it may recognize an epitope on the EGFR that differs from the one to which cetuximab binds.29, 30
Given the complexity of the EGFR signaling network and the technical restraints related to protein detection by IHC, an alternative approach to predict tumor response to anti-EGFR therapy is to analyze the EGFR gene status. Gene amplification, transcriptional upregulation, or abnormal receptor structure secondary to genetic alterations (eg, mutation or polymorphism) could potentially be reasons for EGFR overexpression,28, 31 and therefore predictors of response to anti-EGFR therapy. From the technical point of view, the detection of gene signals would overcome certain restraints related to IHC such as storage time. It has been shown that EGFR staining intensity declines as the storage time of the tissue samples prolongs.30 The preservation of the gene, on the other hand, would be much less of a problem. If the gene is not preserved, no gene signals will be seen in either the tumor cells or nontumor cells, and the case can be identified as having a technical problem. The chance of having a false-negative result in this situation is therefore minimized.
Our data confirm previous observations5 that a very high percentage of colorectal carcinomas (>75%) have immunohistochemical positivity for EGFR. Clinical trials thus far have demonstrated a response rate of only 9–23% to cetuximab in colorectal cancer patients.9, 10 It is evident that a considerable proportion of patients with EGFR IHC-positive tumors will not achieve significant benefit from the use of cetuximab. The frequency of gene amplification, on the other hand, is only 12% in primary colorectal carcinomas and 8% in metastases. The presence of a small but defined proportion of colorectal cancers that show EGFR gene amplification is in agreement with a recent report that used fluorescent in situ hybridization and IHC,32 but differ from earlier studies that used Southern blot- or polymerase chain reaction (PCR)-based methods. In studies that used Southern blotting33 or PCR,34 no gene amplification was demonstrated in colorectal carcinoma cell lines or tissues microdissected from paraffin blocks, despite the presence of protein expression. Such discrepant results may be explained by dilution of tumor lysates by non-neoplastic tissues or sampling limitations since only a small fraction of EGFR IHC-positive primary colorectal carcinomas showed gene amplification as demonstrated by this study and by the study by Ooi et al.32 Using fluorescent in situ hybridization and IHC, Ooi et al32 showed that only 21% (11/53) of EGFR IHC-positive (1+, 2+ or 3+) colorectal carcinomas showed gene amplification.
Our finding that many EGFR-positive colorectal carcinomas as detected by IHC (including 67% of IHC 3+ tumors) do not show gene amplification suggests a poor correlation between EGFR protein expression and gene amplification. Although the few tumors (3/147) with foci of high-level EGFR gene amplification all showed intense immunohistochemical staining in the same foci, the tumors with low-level amplification showed immunohistochemical positivity ranging from 1+ to 3+. The data demonstrate that EGFR expression as measured by IHC does not predict gene status. This contrasts the status of HER2 in colorectal carcinomas in that HER-2 overexpression by IHC is highly correlated with gene amplification.35
Given the fact that EGFR gene amplification is detected in only a small fraction of EGFR IHC-positive colorectal tumors, and similar proportion of patients with EGFR-positive tumors responded to cetuximab-based therapy, it seems plausible to test the paramount question whether EGFR gene amplification is a better predictor of response to anti-EGFR therapy with additional clinical studies. However, our finding that none of the tumors that did not express EGFR by IHC showed gene amplification seems to suggest that gene amplification by chromogenic in situ hybridization may not be a reliable indicator either. This argument arose because of the recent demonstration that a small proportion of EGFR-negative colorectal cancer patients also respond to cetuximab-based treatment.13 The lack of any gene-amplified tumors in the IHC-negative group in our study suggests that chromogenic in situ hybridization would have missed the same fraction of IHC-negative tumor patients who might respond to cetuximab. The number of IHC-negative cases, however, is rather small in our study, as is the number of patients with EGFR-negative tumors that have been studied for response to anti-EGFR therapy. Obviously, a clear answer to the utility of gene amplification in predicting treatment response requires further investigation.
To our knowledge, this is the first study to use chromogenic in situ hybridization for the detection of EGFR gene amplification in colorectal carcinomas. The interpretation of in situ hybridization signals in these tumors requires caution, largely owing to the fact that colorectal carcinoma cell nuclei often overlap and form pseudostratification. High-level amplification is often easily discernible even when the nuclei are crowded. However, low-level amplification with <10 signals per nucleus requires careful signal enumeration. Areas of nonoverlapping nuclei should be chosen for enumeration whenever possible. However, it is sometimes unavoidable to perform signal enumeration in an area of overlapping nuclei. In such instances, it is more accurate to count the total number of nuclei in a small area and the total number of signals in these nuclei, instead of enumerating each nucleus individually. Despite these technical restraints, chromogenic in situ hybridization is generally a simple and practical technique to study gene amplification.
In summary, EGFR gene amplification is much less frequent than protein expression measurable by IHC in colorectal carcinomas. IHC has a low specificity in predicting EGFR gene amplification in these tumors. Further investigation is needed to determine whether EGFR gene amplification bears any informative value in predicting response to anti-EGFR therapy.
References
Ciardiello F, Tortora G . A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res 2001;7:2958–2970.
Ciardiello F, Tortora G . Epidermal growth factor receptor (EGFR) as a target in cancer therapy: understanding the role of receptor expression and other molecular determinants that could influence the response to anti-EGFR drugs. Eur J Cancer 2003;39:1348–1354.
Starling N, Cunningham D . Monoclonal antibodies against vascular endothelial growth factor and epidermal growth factor receptor in advanced colorectal cancers: present and future directions. Curr Opin Oncol 2004;16:385–390.
Baselga J . The EGFR as a target for anticancer therapy—focus on cetuximab. Eur J Cancer 2001;37 (Suppl 4):S16–S22.
Goldstein NS, Armin M . Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer 2001;92:1331–1346.
Nicholson RI, Gee JM, Harper ME . EGFR and cancer prognosis. Eur J Cancer 2001;37 (Suppl 4):S9–S15.
Ciardiello F, Bianco R, Damiano V, et al. Antitumor activity of sequential treatment with topotecan and anti-epidermal growth factor receptor monoclonal antibody C225. Clin Cancer Res 1999;5:909–916.
Saltz LB, Hochster H, Rubin M, et al. Cetuximab (IMC-C225) plus irinotecan (CPT-11) is active in CPT-11-refractory colorectal cancer (CRC) that expresses epidermal growth factor receptor (EGFR). Proceedings of the American Society Clinical Oncologists. San Francisco, CA, 2001 (Abstract).
Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337–345.
Saltz LB, Meropol NJ, Loehrer Sr PJ, et al. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 2004;22:1201–1208.
Iqbal S, Lenz HJ . Integration of novel agents in the treatment of colorectal cancer. Cancer Chemother Pharmacol 2004;54 (Suppl 1):S32–S39.
Lenz H-J, Mayer RJ, Gold PJ, et al. Activity of cetuximab in patients with colorectal cancer refractory to both irinotecan and oxaliplatin. Proceedings of the American Society Clinical Oncologists. New Orleans, LA, 2004.
Chung K, Shia J, Kemeny NE, et al. Ectuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803–1810.
Isola J, Tanner M, Forsyth A, et al. Interlaboratory comparison of HER-2 oncogene amplification as detected by chromogenic and fluorescence in situ hybridization. Clin Cancer Res 2004;10:4793–4798.
Isola J, Tanner M . Chromogenic in situ hybridization in tumor pathology. Methods Mol Med 2004;97:133–144.
Bhargava R, Lal P, Chen B . Chromogenic in situ hybridization for the detection of HER-2 gene amplification in breast cancer with an emphasis on tumors with borderline and low-level amplification: does it measure up to fluorescence in situ hybridization? Am J Clin Pathol 2005;123:237–243.
Madrid MA, Lo RW . Chromogenic in situ hybridization (CISH): a novel alternative in screening archival breast cancer tissue samples for HER-2/neu status. Breast Cancer Res 2004;6:R593–R600.
Marquez A, Wu R, Zhao J, et al. Evaluation of epidermal growth factor receptor (EGFR) by chromogenic in situ hybridization (CISH) and immunohistochemistry (IHC) in archival gliomas using bright-field microscopy. Diagn Mol Pathol 2004;13:1–8.
Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847.
Henshall S . Tissue microarrays. J Mammary Gland Biol Neoplasia 2003;8:347–358.
Horvath L, Henshall S . The application of tissue microarrays to cancer research. Pathology 2001;33:125–129.
Hoos A, Urist MJ, Stojadinovic A, et al. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol 2001;158:1245–1251.
Shergill IS, Shergill NK, Arya M, et al. Tissue microarrays: a current medical research tool. Curr Med Res Opin 2004;20:707–712.
Camp RL, Charette LA, Rimm DL . Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000;80:1943–1949.
Diaz LK, Gupta R, Kidwai N, et al. The use of TMA for interlaboratory validation of FISH testing for detection of HER2 gene amplification in breast cancer. J Histochem Cytochem 2004;52:501–507.
Bhargava R, Lal P, Chen B . Feasibility of using tissue microarrays for the assessment of HER-2 gene amplification by fluorescence in situ hybridization in breast carcinoma. Diagn Mol Pathol 2004;13:213–216.
Pirie W . Konchheere Tests for Ordered Alternatives. John Wiley & Sons, Inc.: New York, 1983, pp 315–318.
Mendelsohn J, Baselga J . Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol 2003;21:2787–2799.
Erlichman C, Sargent DJ . New treatment options for colorectal cancer. N Engl J Med 2004;351:391–392.
Atkins D, Reiffen KA, Tegtmeier CL, et al. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem 2004;52:893–901.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–2139.
Ooi A, Takehana T, Li X, et al. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol 2004;17:895–904.
Radinsky R, Risin S, Fan D, et al. Level and function of epidermal growth factor receptor predict the metastatic potential of human colon carcinoma cells. Clin Cancer Res 1995;1:19–31.
Layfield LJ, Bernard PS, Goldstein NS . Color multiplex polymerase chain reaction for quantitative analysis of epidermal growth factor receptor genes in colorectal adenocarcinoma. J Surg Oncol 2003;83:227–231.
Nathanson DR, Culliford AT, Shia J, et al. HER 2/neu expression and gene amplification in colon cancer. Int J Cancer 2003;105:796–802.
Acknowledgements
This work was supported in part by NCI 2 P01 CA65930-05A2.
Author information
Authors and Affiliations
Corresponding author
Additional information
Presented at the 94th Annual Meeting of the United States and Canadian Academy of Pathology, San Antonio, TX, February 26–March 4, 2005.
Rights and permissions
About this article
Cite this article
Shia, J., Klimstra, D., Li, A. et al. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study. Mod Pathol 18, 1350–1356 (2005). https://doi.org/10.1038/modpathol.3800417
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800417
Keywords
This article is cited by
-
Adaptive resistance to lorlatinib via EGFR signaling in ALK-rearranged lung cancer
npj Precision Oncology (2023)
-
Depleting receptor tyrosine kinases EGFR and HER2 overcomes resistance to EGFR inhibitors in colorectal cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer
Biological Research (2019)
-
EGFR Gene Amplification and KRAS Mutation Predict Response to Combination Targeted Therapy in Metastatic Colorectal Cancer
Pathology & Oncology Research (2017)
-
Oncolytic Activity of a Recombinant Measles Virus, Blind to Signaling Lymphocyte Activation Molecule, Against Colorectal Cancer Cells
Scientific Reports (2016)