Abstract
Secretory carcinomas of the breast are rare tumors with distinct histologic features, recurrent t(12;15)(p13;q25) translocation resulting in ETV6–NTRK3 gene fusion and indolent clinical behavior. Mammary analog secretory carcinomas arising in other sites are histopathologically similar to the breast tumors and also harbor ETV6–NTRK3 fusions. Breast secretory carcinomas are often triple (estrogen and progesterone receptor, HER2) negative with a basal-like immunophenotype. However, genomic studies are lacking, and whether these tumors share genetic features with other basal and/or triple negative breast cancers is unknown. Aside from shared ETV6–NTRK3 fusions, the genetic relatedness of secretory carcinomas arising in different sites is also uncertain. We immunoprofiled and sequenced 510 cancer-related genes in nine breast secretory carcinomas and six salivary gland mammary analog secretory carcinomas. Immunoprofiles of breast and salivary gland secretory carcinomas were similar. All the tumors showed strong diffuse MUC4 expression (n=15), and SOX10 was positive in all nine breast and in five out of six salivary gland tumors. All breast secretory carcinomas were triple negative or weakly ER-positive, and all tumors at both the sites expressed CK5/6 and/or EGFR, consistent with a basal-like phenotype. Sequencing revealed classic ETV6–NTRK3 fusion genes in all cases, including in carcinoma in situ of one breast tumor. Translocations were reciprocal and balanced in six out of nine breast and three out of six salivary gland tumors and were complex in three others. In contrast to most breast basal carcinomas, the mutational burden of secretory carcinomas was very low, and no additional pathogenic aberrations were identified in genes typically mutated in breast cancer. Five (56%) breast and two (33%) salivary gland tumors had simple genomes without copy number changes; the remainder had very few changes, averaging 1.3 per tumor. The ETV6–NTRK3 derivative chromosome was duplicated in one breast and one salivary gland tumor, and was the only copy number change in the latter. The findings highlight breast secretory carcinoma as a subtype more closely related to mammary analog secretory carcinoma than to basal/triple negative breast cancers of no special type. Lack of pathogenic mutations in common cancer-related genes suggests that ETV6–NTRK3 alone may suffice to drive these tumors and likely helps explain their indolent behavior.
Similar content being viewed by others
Main
Secretory carcinomas of the breast are a very rare special subtype of breast cancer, estimated to comprise approximately 0.15% of all breast carcinomas, with distinctive histopathologic features and favorable clinical behavior.1, 2, 3, 4, 5, 6 Initially described in children and adolescents, more recent studies indicate that secretory carcinomas are more common in adults.4, 5, 7, 8 The overall prognosis is favorable even in the presence of local lymph node metastasis, with only vanishingly rare distant metastases. Five- and 10-year disease-related survival is 94% and 91%, respectively.4, 8
Secretory carcinomas are prototypical of genotype–phenotype correlation among tumors, demonstrating characteristic histologic and immunohistochemical features and a recurrent t(12;15)(p13;q25) translocation that creates an ETV6–NTRK3 fusion gene, which is specific among breast tumors.1, 2, 5, 9, 10 ETV6–NTRK3 encodes a chimeric tyrosine kinase that signals through Ras-mitogen activated protein kinase and phosphatidyl inositol-3 kinase pathways to transform cells of multiple lineages.11, 12, 13, 14, 15, 16 Morphologic features of breast secretory carcinomas are those of grade 1 or grade 2 tumors comprised of polygonal cells with eosinophilic or vacuolated cytoplasm and small, round to oval nuclei arranged in microcystic, solid, tubular, or papillary growth patterns and associated with periodic acid Schiff (PAS)-positive secretions.1, 2, 5, 6, 17 High-grade tumors have only rarely been reported.1 The tumor cells characteristically express S100 protein, mammaglobin, and STAT5a and are either triple (estrogen receptor [ER], progesterone receptor [PR], and HER2) negative or only weakly ER/PR-positive.1, 2, 17, 18, 19, 20 Several studies have demonstrated a basal-like phenotype of secretory carcinomas by immunohistochemistry, including positivity for EGFR and the basal keratin CK5/6, in addition to the triple negative or weakly ER-positive status.1, 17 However, the excellent prognosis and long-term outcome of patients with secretory carcinoma, even in the presence of axillary lymph node metastasis, is in stark contrast to the majority of triple negative or basal breast cancers, highlighting the heterogeneity of basal tumors and the need for comprehensive histopathologic evaluation.4, 8, 21, 22, 23 Although large sequencing projects, including The Cancer Genome Atlas (TCGA) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), have interrogated the genomic landscape of breast cancers, these studies have focused on the overwhelming majority of invasive carcinomas of no special type and have generally not included special histologic subtypes.21, 22, 24, 25, 26, 27, 28, 29, 30, 31 Indeed, aside from targeted studies of the ETV6–NTRK3 fusion and array comparative hybridization studies, the genomics of secretory carcinomas remain largely unexplored, and next-generation DNA sequencing studies of these rare tumors are lacking.1, 2, 9, 17, 32, 33 Accordingly, it remains unknown whether secretory carcinomas harbor genomic hallmarks of other breast cancers, especially basal-like and/or triple negative tumors, to suggest a genetic relatedness.
Recent studies have identified mammary analog secretory carcinomas at other sites, including major and minor salivary glands, thyroid and skin, which show similar morphologic and immunophenotypic features to secretory carcinomas of the breast and also harbor ETV6–NTRK3 fusions.34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44 Indeed, a number of salivary-like tumors may arise in the breast, suggesting a histogenetic relationship of these tumors irrespective of the site of origin. These include acinic cell carcinomas, mucoepidermoid carcinomas, and adenoid cystic carcinomas, among other even rarer tumors.45, 46, 47, 48, 49, 50, 51 Accordingly, special breast cancer subtypes may be more closely related to their respective counterparts at other sites than to other primary breast carcinomas, but supportive genetic data are sparse and may be tumor-dependent. A recent study suggested that adenoid cystic carcinomas arising in the breast lack TP53 and PIK3CA mutations typical of other basal-like breast cancers and share a variety of mutated pathways with salivary gland adenoid cystic carcinomas, including but not limited to the characteristic MYB–NFIB gene fusion, suggesting that the breast tumors are more similar to their salivary gland counterparts than they are to other breast cancers.45 The genetic data do not address the discordance in clinical behavior between the tumors. On the other hand, acinic cell carcinomas of the breast harbor hallmark genetic alterations of other typical triple negative breast carcinomas, including frequent TP53 mutations and complex patterns of copy number alterations, not present in salivary gland acinic cell carcinomas, suggesting that the two tumors are not related despite histologic similarity.52, 53 Given the lack of genomic data or comparative genetic studies, the relatedness of secretory carcinomas of the breast to analog tumors arising in other sites remains unknown.
In this study, we used capture-based next-generation sequencing of 510 cancer-related genes to more comprehensively characterize the genomics of breast secretory carcinomas and to compare the genetics of these rare tumors with those of mammary analog secretory carcinomas and with published data of other breast cancers, including triple negative and/or basal tumors. The findings shed light on our understanding of secretory carcinoma biology and may help explain the favorable clinical behavior of these tumors.
Materials and methods
Study Population
This study was approved by the institutional review boards of the University of California San Francisco, Kaiser Permanente and Weill Cornell. Nine breast secretory carcinomas and six mammary analog secretory carcinomas of the salivary gland were identified in the consultation services and Pathology department archives of our respective institutions, spanning years 2006 to 2015 (SC1-9 and MASC1-6, respectively). All the specimens were fixed in 10% neutral buffered formalin and embedded in paraffin. Clinical information was obtained from the electronic medical records of our respective institutions and/or from the referring facilities.
Capture-Based Next-Generation DNA Sequencing
Matched normal and tumor tissues were selected from the nine breast secretory carcinomas and six salivary gland mammary analog secretory carcinomas for capture-based next-generation DNA sequencing. Sequencing was performed at the UCSF Clinical Cancer Genomics Laboratory, using an assay that targets the coding regions of 510 cancer-related genes, select introns from 42 genes, and TERT promoter with a total sequencing footprint of 2.8 Mb (UCSF500 panel; Supplementary Table S1). Sequencing libraries were prepared from genomic DNA extracted from both tumor and normal formalin-fixed paraffin-embedded tissue. For one breast secretory carcinoma, DNA from associated ductal carcinoma in situ was extracted and analyzed separately. Target enrichment was performed by hybrid capture using a custom oligonucleotide library. Sequencing was performed on an Illumina HiSeq 2500. Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using the following software packages: BWA: 0.7.13, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: Appistry v2015.1.1–3.4.46-0-ga8e1d99, CNVkit: 0.7.2, Pindel: 0.2.5b8, SATK: Appistry v2015.1.1-1-gea45d62, Annovar: v2016Feb01, Freebayes: 0.9.20 and Delly: 0.7.2.54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64 Only insertions/deletions up to 100 base pairs in length were included in the mutational analysis. Somatic single-nucleotide variants and insertions/deletions were visualized and verified using Integrated Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and Nexus Copy Number (Biodiscovery, Hawthorne, CA, USA).60 Large-scale chromosomal changes were defined as those involving entire chromosomes or chromosome arms.
Targeted RNA Sequencing
Targeted RNA sequencing was performed on one mammary analog secretory carcinoma without an identified fusion gene by DNA sequencing (MASC5). RNA was isolated from 5 micron thick unstained formalin-fixed paraffin-embedded tumor sections on glass slides using Qiagen RNeasy kit according to the manufacturer’s instructions (Qiagen, Germantown, MD, USA). Complementary DNA was subsequently prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina according the manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA). Sequencing and analysis was performed using the UCSF500 platform as described above.
Fluorescence In Situ Hybridization
For ETV6 fluorescence in situ hybridization, 5 micron thick unstained formalin-fixed paraffin-embedded sections on glass slides were baked at 60 °C for 1 h, rinsed in 100% ethanol, then pretreated with 0.2 N HCl (20 min, room temperature), followed by 1 M NaSCN (30 min, 80 °C) before protease digestion with pepsin (2.5 mg/ml pepsin for 27 min at 37 °C). The slides were then fixed in 10% phosphate-buffered saline-buffered formalin, rinsed, dehydrated in an ethanol series, and air dried. The slides were hybridized overnight at 37 °C with an equimolar mixture of Vysis LSI ETV6 (Tel) SpectrumOrange and (Cen) SpectrumGreen DNA probes (07J77-003 and 07J77-004, respectively; Abbott Molecular, Des Plaines, IL, USA) after denaturing for 5 min at 80 °C. The slides were washed to remove unbound probes and counterstained with 4,6 diamidino-2-phenylidole. Enumeration of the fusion and break-apart signals was conducted using a Zeiss fluorescence microscope.
Tissue Microarray Construction
Tissue microarrays were created from ER-/PR-/HER2- (n=111), ER+/HER2- (n=32), and HER2+ (n=30) untreated breast cancers diagnosed at UCSF from 1997 to 2009.65 All the specimens were fixed in 10% buffered formalin and embedded in paraffin. Three 1 mm punch biopsy tissue cores were obtained from each tumor to create triplicates for analysis. Only tumors with at least two evaluable cores with tumor tissue were used for analysis.
Immunohistochemistry
Immunohistochemistry was performed on whole tissue sections (all antibodies) or tissue microarrays (MUC4 and SOX10). The following antibodies were used: S100 (polyclonal, 1:2000, DAKO, Carpinteria, CA, USA), SOX10 (EP268, 1:250, Cell Marque, Rocklin, CA, USA), MUC4 (8G7, 1:500, Millipore, Billerica, MA, USA), GATA3 (L50-823, 1:50, Biocare Medical, Concord, CA, USA), mammaglobin (304-1A5, 1:4, DAKO), gross cystic disease fluid protein 15 (GCDFP-15; 23A3, undiluted, Covance, Dedham, MA, USA), CK5/6 (D5/16B4, 1:200 with anti-background, Millipore), and EGFR (5B7, undiluted, Roche, Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval was as follows: for S100—none; for SOX10—Bond epitope retrieval solution 2 (Leica Biosystems, Buffalo Grove, IL, USA); for MUC4—Bond epitope retrieval solution 1 (Leica Biosystems); for GATA3—Bond epitope retrieval solution 2 (Leica Biosystems); for mammaglobin and GCDFP-15—Bond epitope retrieval solution 1 (Leica Biosystems); for CK5/6—Bond epitope retrieval solution 1 (Leica Biosystems); and for EGFR, CC1 (Roche, Ventana Medical Systems). For a subset of secretory carcinomas received in consultation, immunohistochemistry for some markers was performed at the referring institution.
For estrogen receptor, progesterone receptor, and HER2, positive staining was defined according to ASCO/CAP guidelines.66, 67 For all other markers, positive expression was defined as any cytoplasmic (MUC4, mammaglobin, GCDFP-15, CK5/6), nuclear (SOX10, GATA3), combined cytoplasmic/nuclear (S100), or combined membranous/cytoplasmic (EGFR) staining. Percentages of tumor cells stained by each antibody were recorded, with diffuse staining defined as >80% of tumor cell staining. For invasive ductal carcinomas with >80% of MUC4 tumor cell staining in all evaluable cores on tissue microarrays, whole tissue sections were subsequently stained.
Results
Clinicopathologic Features of Secretory Carcinomas
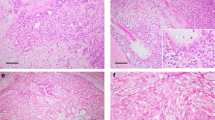
Clinicopathologic features of the breast secretory carcinomas analyzed in this study are shown in Table 1 and Figure 1. Patients included eight women and one man; ages ranged from 13 to 71 years (mean 43, median 45). All tumors showed characteristic histologic features of secretory carcinoma, including polygonal cells with eosinophilic or vacuolated cytoplasm and grade 2 nuclei, growing in pure or mixed combinations of syncytial microcystic, solid, papillary, or tubular patterns and associated with dense eosinophilic secretions (Figure 1). Modified Scarff-Bloom-Richardson grade was 1 in all cases, with sizes ranging from 0.5 to 2.5 cm (mean size 1.2 cm; Table 1). Associated carcinoma in situ was present in six (67%) cases. Four (50%) tumors arose in the subareolar region, and two (25%) were present in axillary ectopic breast tissue (n=8). One tumor (SC4) arising in the axilla also demonstrated tumor cells with prominent apocrine cytologic features (eosinophilic cytoplasmic granules and rounded nuclei with prominent nucleoli) in many areas, intimately admixed with tumor cells showing characteristic secretory features (Figure 1). Two patients (SC1 and SC2) presented with single, small ipsilateral axillary lymph node metastases. Clinical follow-up was available for five patients, none of which showed recurrences or death due to breast cancer (mean follow-up interval 78 months, range 45–98 months; Table 1).
Representative images of breast secretory carcinomas and salivary gland mammary analog secretory carcinomas. Breast secretory carcinomas are shown in (a–i) and salivary gland mammary analog secretory carcinomas are shown in (j–o). Most tumors showed variable combinations of microcystic, solid, papillary and/or tubular growth, but only selected areas are depicted here. (a and b) SC1, showing characteristic microcystic growth pattern (in a) and foci of tubular infiltration (in b), each associated with eosinophilic luminal secretions. (c) This tumor (SC2) demonstrated microcystic areas with prominent cytoplasmic vacuolization, reminiscent of pseudolactational change. (d) Microcystic change (right) merging with more solid growth (left) in SC3. (e) This tumor (SC4) arose within ectopic axillary breast tissue and showed tumor cells with apocrine features, including prominent eosinophilic granules and rounded nuclei with large nucleoli, admixed with more typical secretory carcinoma cells. The growth pattern was predominantly microcystic (shown) and focally papillary. (f) This tumor showed well-developed papillary features, admixed with superimposed microcystic growth (SC5). (g) SC6, showing solid and microcystic growth. (h) Occasionally, the syncytial microcystic growth pattern of secretory carcinomas may mimic fenestrated growth of florid usual ductal hyperplasia (SC7). (i) SC9, with typical intermediate grade nuclei and eosinophilic luminal secretions. (j) MASC1, (k) MASC2, with nuclear features akin to grade 1 tumors in the breast. (l) This tumor (MASC3) showed characteristic microcystic and papillary growth in many areas and focally grew as distinctly infiltrative cords within desmoplastic-appearing stroma. (m) Similar to SC2, this mammary analog secretory carcinoma (MASC4) demonstrated prominent cytoplasmic vacuolization and nuclear hobnailing, resembling pseudolactational change of the breast, in this focus superimposed on papillary fibrovascular cores. (n) This tumor (MASC5) showed infiltrative nests and cords with microcystic change and focal papillary growth (top left). (o) The only salivary gland tumor in this study not arising in the parotid gland, MASC6, showed almost exclusively solid infiltrative growth, as shown here. All images hematoxylin and eosin.
Salivary gland mammary analog secretory carcinomas demonstrated similar cytologic and architectural features as the breast tumors (Figure 1 and Table 1). Five (83%) tumors arose in the parotid gland and one (17%) arose in a minor buccal salivary gland. No patients showed recurrences, and all are alive without evidence of disease (mean follow-up interval 29 months, range 9–47 months; Table 1).
Immunohistochemical Profiles of Secretory Carcinomas
Breast secretory carcinomas and mammary analog secretory carcinomas showed similar immunophenotypes (Table 2). Five (56%) breast secretory carcinomas were triple negative, and all four ER-positive tumors showed weak to moderate expression in only ~2 to 15% of tumor cells. As expected, all breast (n=7) and salivary gland (n=6) carcinomas expressed S100 protein and mammaglobin, with diffuse expression identified in most tumors. In addition, diffuse and strong MUC4 expression was identified in all 15 secretory carcinomas regardless of site, and SOX10 was positive in all nine breast and five out of six salivary gland tumors (Figure 2 and Table 2). In comparison with breast secretory carcinomas, only 3 (2%) of 144 invasive ductal carcinomas of no special type demonstrated strong diffuse MUC4 expression (P<0.001 vs breast secretory carcinomas), including 2 (2.4%) of 82 high-grade triple negative tumors (P<0.001) and 1 (3.1%) of 32 ER+/HER2− tumors (P<0.001); no (0%) HER2+ tumors were diffusely positive for MUC4 (n=30; P<0.001). Twelve of 93 (13%) triple negative invasive ductal carcinoma of no special type diffusely expressed SOX10 (P<0.001 vs breast secretory carcinomas), one of which was also diffusely positive for MUC4.
Immunohistochemical profiles of representative secretory carcinomas. (a–c) Breast secretory carcinomas and (d–f) salivary gland mammary analog secretory carcinomas express CK5/6 (b and e) and/or EGFR (c and f); SC2 and MASC3 shown, respectively, × 400. (g–i) Breast secretory carcinomas and (j–l) salivary gland mammary analog secretory carcinomas demonstrate strong diffuse MUC4 staining (h and k) and diffuse SOX10 expression (i and l); SC3 and MASC1 shown, respectively, × 400.
All secretory carcinomas expressed cytokeratin CK5/6 (n=8 breast, n=5 salivary gland) and/or EGFR (n=5 breast, n=5 salivary gland), consistent with a basal-like immunophenotype previously reported for breast secretory carcinomas1, 17 (Table 2 and Figure 2). EGFR tended to be positive in a higher proportion of tumor cells than CK5/6, which was only focally expressed (≤5% of tumor cells) in five out of eight (63%) breast and three out of five (60%) salivary gland tumors. In contrast, EGFR was expressed in ≥20% of tumor cells in three out of five (60%) breast and all five (100%) salivary gland tumors (Figure 2 and Table 2). All tumors that were stained for both markers showed positivity for either CK5/6 or EGFR in ≥10% of tumor cells (Table 2).
Genomics of Breast Secretory Carcinomas with Comparison to Salivary Gland Mammary Analog Secretory Carcinomas
All tested breast (n=9) and salivary gland analog (n=5) secretory carcinomas were positive for ETV6 gene rearrangement by break-apart fluorescence in situ hybridization probe, including the associated carcinoma in situ component of one breast secretory carcinoma case (SC5; Figure 3).
Genomic features of secretory carcinomas. (a) Summary of fluorescence in situ hybridization (FISH) and next-generation sequencing results for all 15 secretory carcinomas. All cases demonstrated ETV6 gene break-apart by FISH. All cases demonstrated ETV6–NTRK3 gene fusion by DNA sequencing except for MASC5, in which the ETV6–NTRK3 fusion was not detected by DNA sequencing but was detected by RNA sequencing. The oncogenic derivative chromosome 15 (der(15)) was duplicated in two cases (SC3 and MASC2). All cases had simplex genomes with 0–2 chromosomal gains and 0–2 chromosomal losses per case. None of the cases had focal amplifications or deep deletions. None of the cases demonstrated additional pathogenic mutations. Several cases had one or two somatic variants of uncertain significance (VUS). (b) In all cases, the ETV6–NTRK3 fusion occurred with DNA breakpoints in intron 5 of ETV6 (NM_001987) and intron 14 of NTRK3 (NM_001012338), resulting in a predicted fusion gene composed of N-terminal exons 1–5 of ETV6, which encompass the sterile alpha motif (SAM) dimerization domain, and C-terminal exons 15–20 of NTRK3, which includes the protein kinase domain. (c and d) ETV6 break-apart FISH images. Orange, green, and yellow dots represent 5′ ETV6 probe (telomeric), 3′ ETV6 probe (centromeric), and fused probes (normal), respectively. Cases with copy neutral ETV6–NTRK3 gene fusions (c) had one fused signal and single separate orange and green signals representing a break-apart. Cases SC3 and MASC2 (d) had one fused signal but two separate orange signals (representing the duplicated oncogenic derivative chromosome) and a single separate green signal.
The mean target coverage of next-generation DNA sequencing was 459 unique reads per target interval for breast secretory carcinomas (range 154–880) and 512 for salivary gland mammary analog secretory carcinomas (range 317–699; Table 3 and Supplementary Table S2). All nine breast secretory carcinomas (n=9) and five of six salivary gland mammary analog secretory carcinomas revealed ETV6–NTRK3 gene fusions by DNA sequencing (Figure 3). In all cases, the DNA breakpoints were in intron 5 of ETV6 (NM_001987) and intron 14 of NTRK3, (NM_001012338), resulting in a predicted fusion gene composed of N-terminal exons 1–5 of ETV6, which encompass the sterile alpha motif (SAM) dimerization domain, and C-terminal exons 15–20 of NTRK3, which includes the protein kinase domain (Figure 3b).9 One salivary gland tumor (MASC5) did not demonstrate an ETV6–NTRK3 gene fusion by DNA sequencing, despite a mean sequencing coverage of 536 reads per interval in this case. Subsequent targeted RNA sequencing of MASC5 did reveal the same ETV6–NTRK3 fusion (exons 1–5 of ETV6 fused to exons 15–20 of NTRK3) as in the other cases (Figure 3b); thus, the ETV6 breakpoint in this case likely occurred in a small region of ETV6 intron 5 that is poorly covered in all of the samples. In one out of nine breast and two of six salivary gland tumors, the rearrangements were complex and involved additional genes (Figure 3 and Table 3), whereas balanced reciprocal t(12;15)(p13;q25) translocations were identified in six out of nine breast and three of six salivary gland tumors (Figure 3 and Table 3). This translocation results in a derivative chromosome 15, which contains the oncogenic fusion. Two of the cases with reciprocal fusions (SC3 and MASC2) demonstrated duplication of the oncogenic derivative chromosome (Figure 3a and d). Both cases demonstrated copy gain of the distal portion of chromosome 12p extending from intron 5 of ETV6 gene to the telomere (Figure 4c,Supplementary Figure S1), which correlates with gain of the 5′ ETV6 FISH probe (orange signal, Figure 3d). In MASC2, the distal 12p gain is accompanied by a copy gain in 15q extending from intron 14 of NTRK3 to the centromere (Figure 4c, Supplementary Figure S1), which is the expected result if the oncogenic derivative chromosome is duplicated. In SC3, the distal 12p gain is accompanied by a copy loss in 15q from intron 14 of NTRK3 to the telomere (Figure 4c and Supplementary Figure S1), which is the expected result if the oncogenic derivative chromosome is duplicated and the normal copy of chromosome 15 is lost.
Copy number profiles of selected secretory carcinomas. (a) Copy number analysis reveals lack of copy number alterations in many secretory carcinomas. Representative breast secretory carcinomas (SC5 and SC6) and a salivary gland mammary analog secretory carcinoma (MASC3) are shown. (b) Secretory carcinomas with copy number changes demonstrate only a few chromosomal gains or losses per tumor. Representative breast secretory carcinomas (SC2 and SC4) and a salivary gland mammary analog secretory carcinoma (MASC1) harbor recurrent copy gain of chromosome 16 (black arrows) and very few additional chromosomal changes. (c) Duplication of the oncogenic rearranged derivative chromosome 15 harboring ETV6–NTRK3 in one breast (SC3) and one salivary gland (MASC2) tumor. The copy number profile of MASC2 (bottom) shows copy gain of distal 12p from the ETV6 breakpoint to the telomere (black arrow), as well as copy gain of 15q from the NTRK3 breakpoint and extending proximally to the centromere (red arrow). The findings are indicative of duplication of the derivative chromosome harboring the ETV6–NTRK3 fusion, which is the only copy number alteration in this tumor. The copy number profile of SC3 (top) also shows copy gain of distal 12p from the ETV6 breakpoint to the telomere (black arrow), which in this case is associated with copy neutral 15q from the centromere to the NTRK3 breakpoint and copy loss of distal 15q from the NTRK3 breakpoint to the telomere (red arrow). The findings are consistent with duplication of the derivative chromosome harboring the ETV6–NTRK3 fusion plus loss of normal chromosome 15, which is also supported by allelic imbalance of copy neutral proximal 15q by single-nucleotide polymorphism analysis and fluorescence in situ hybridization (not shown here; see also text and Supplementary Figure S1). Red arrows represent large-scale (chromosome or chromosome arm level) copy number alterations private to only one tumor.
The mutational burden over the 2.8 megabase footprint of the panel was very low in both groups, and there were from 0 to 2 nonsynonymous coding mutations per tumor and averaging 0.7 and 0.8 variants per breast and salivary gland tumor, respectively (Table 3 and Figure 3). No mutations were identified in genes frequently mutated in breast cancers, including basal carcinomas.22, 25, 26, 27, 28, 29 Indeed, no known pathogenic mutations were identified in any of the cancer-related genes on the panel, nor were recurrently mutated genes found. The few identified variants of unknown significance are shown in Table 3. Four of the breast (44%) and two of the salivary gland (33%) carcinomas did not harbor any somatic coding mutations, including one of the breast tumors with lymph node metastasis (SC2; Table 3).
Copy number analysis revealed simple genomes without any chromosomal gains or losses in five (56%) breast secretory carcinomas and two (33%) mammary analog secretory carcinomas. In one additional salivary gland tumor, the only copy number change was duplication of the rearranged derivative chromosome 15 with ETV6–NTRK3 gene fusion (MASC2; Table 3, Figure 3 and Supplementary Figure S1). The remainder of the tumors in both the groups showed only very few copy number changes, ranging from 1 to 2 gains or losses per tumor. The average number of copy number changes per tumor was 1.3 (0.9 in breast secretory carcinomas and 2 in salivary gland analog secretory carcinomas). Recurrent gains included distal chromosome 12p (part of the rearranged oncogenic derivative chromosome harboring ETV6–NTRK3) in SC3 and MASC2, chromosome 8q (two breast tumors) and chromosome 16 (two breast and one salivary gland tumor); chromosome 22 was lost in two salivary gland tumors. None of the tumors showed 1q gain or 16q loss characteristic of the low-grade neoplasia pathway in the breast.68 No focal amplifications or deep deletions were identified (Table 3 and Figure 4).
Discussion
This is the first study to analyze the genomics of secretory carcinomas using next-generation sequencing of a large panel of cancer-related genes. In summary, we have shown that secretory carcinomas of the breast and mammary analog secretory carcinomas of the salivary gland have overlapping immunoprofiles, simple genomes with no or few copy number alterations, a very low mutation burden and lack of additional known pathogenic genetic aberrations in 510 common cancer genes aside from ETV6–NTRK3 fusion genes. The findings suggest that, despite a basal-like immunophenotype, secretory carcinomas of the breast do not harbor genomic alterations typical of basal-like carcinomas of no special type and are genetically more similar to mammary analog secretory carcinomas of the salivary gland than they are to other primary breast cancers.
ETV6–NTRK3, initially described in congenital fibrosarcoma, has been identified in tumors of various lineages, including cellular mesoblastic nephroma, acute myeloid leukemia, breast secretory carcinoma, and secretory analog carcinomas of various sites, most frequently of the salivary glands.1, 9, 34, 40, 42, 43, 69, 70, 71, 72, 73, 74, 75, 76 The rearrangement fuses the N-terminal dimerization domain of ETV6 with the C-terminal tyrosine kinase domain of NTRK3, producing a chimeric tyrosine kinase with transforming activity that is activated upon oligomerization.9, 77, 78 By RNA analysis, the most frequent fusion breakpoints are between exon 5 of ETV6 and exon 15 of NTRK3, although variant fusions involving the first four exons of ETV6 and/or exon 14 of NTRK3 have been described in mammary analog secretory carcinomas and radiation-induced thyroid carcinomas.9, 43, 44, 71 Putative alternate ETV6 rearrangements involving unidentified partners have also been described in the salivary gland and have been suggested to demonstrate more aggressive histologic features.43, 79 All breast and salivary gland secretory carcinomas in our study harbored predicted classic ETV6–NTRK3 fusions between ETV6 exon 5 and NTRK3 exon 15. If alternate fusions exist in breast secretory carcinomas, they are likely to be exceedingly rare. One breast and one salivary gland tumor in our series each showed duplication of the entire rearranged chromosome harboring ETV6–NTRK3, and similar findings have been previously reported in breast secretory carcinomas. Of 20 total breast secretory carcinomas in our study and two prior reports,1, 32 duplicated chromosomes harboring ETV6–NTRK3 were identified in 20% of the tumors, in addition to the duplication present in one of the mammary analog secretory carcinomas in our study. Given the apparent frequency of the duplication, it is likely that increased copies of ETV6–NTRK3 are selected for and provide additional advantage to the tumor cells beyond that conferred by the fusion gene alone.
ETV6–NTRK3 transforms multiple cell lineages, and expression causes secretory carcinogenesis in mouse models, but whether the fusion gene is the sole driver or requires additional alterations for tumorigenesis is unclear.9, 11, 12, 16, 77 In this study, we did not identify any additional recurrent alterations using our panel of 510 cancer genes; however, it remains possible that either our panel size or cohort size were too small to detect additional recurrent alterations. Additional sequencing studies of larger numbers of these rare tumors are required to determine whether ETV6–NTRK3 gene fusion is the only driving alteration in secretory carcinoma development or growth.
Although breast secretory carcinomas have a basal-like immunoprofile, their genomics and relationship to other triple negative basal carcinomas of no special type remained unknown before our study. We have shown that secretory carcinomas have a very low mutation burden without pathogenic mutations in genes frequently mutated in basal carcinomas, such as TP53 among others. Furthermore, these tumors have very simple genomes with no or few copy number changes, also in stark contrast to most basal carcinomas of no special type.21 The findings highlight the heterogeneity of basal-like cancers and the importance of histopathology in diagnosis. Identification of a true basal molecular phenotype using gene expression profiling may be useful to determine whether the immunohistochemical profile is a reliable surrogate for basal-like specification in these tumors.80 The absence of chromosomal copy number changes typical of the low-grade neoplasia pathway (such as 1q gain and 16q loss) also further distinguishes these low-grade tumors from breast cancers in the low-grade neoplasia pathway.81 We speculate that the simple genomic profiles of secretory carcinomas may well help explain their favorable clinical course, which is in stark contrast to most other basal breast cancers. Of note, we found no evidence of more aggressive genomic features in breast secretory carcinomas that presented with axillary lymph node metastasis compared with those that did not. This may help explain the notion that even locally metastatic breast secretory carcinomas have favorable outcomes with high long-term survival.4, 8 Unfortunately, insufficient metastatic tumor tissue was available to separately analyze the genomics of the metastases for possible genomic progression. Given our relatively small sample size, additional studies of larger cohorts and of metastatic secretory carcinomas may be instructive.
Our results suggest that breast secretory carcinomas are more closely related to their salivary gland counterparts than they are to other breast cancers. The immunoprofiles of secretory carcinomas and mammary analog secretory carcinomas are nearly identical. In addition to S100 protein, mammaglobin, and Stat5a,17, 20, 36, 39, 42, 82, 83 we show that the breast tumors express MUC4 and SOX10, previously described in their salivary gland counterparts,42, 82, 84 and suggest that these markers may provide an additional diagnostic tool useful in the differential diagnosis to trigger genetic testing. Conversely, mammary analog secretory carcinomas, like the breast tumors, frequently express EGFR and CK5/6 in addition to previously described markers.1, 17, 42, 82 By DNA sequencing, we found no measurable differences in the genomics of secretory carcinomas of either breast or salivary gland. In conjunction with recent studies of adenoid cystic carcinomas of the breast,45 the findings indicate that the rare fusion gene-driven special breast carcinoma subtypes genetically resemble their counterparts at other sites rather than other breast cancers, and that the classification of tumor type is site independent.
References
Del Castillo M, Chibon F, Arnould L et al, Secretory breast carcinoma: a histopathologic and genomic spectrum characterized by a joint specific ETV6-NTRK3 gene fusion. Am J Surg Pathol 2015;39:1458–1467.
Diallo R, Schaefer KL, Bankfalvi A et al, Secretory carcinoma of the breast: a distinct variant of invasive ductal carcinoma assessed by comparative genomic hybridization and immunohistochemistry. Hum Pathol 2003;34:1299–1305.
Din NU, Idrees R, Fatima S et al, Secretory carcinoma of breast: clinicopathologic study of 8 cases. Ann Diagn Pathol 2013;17:54–57.
Jacob JD, Hodge C, Franko J et al, Rare breast cancer: 246 invasive secretory carcinomas from the National Cancer Data Base. J Surg Oncol 2016;113:721–725.
Tavassoli FA, Norris HJ . Secretory carcinoma of the breast. Cancer 1980;45:2404–2413.
Eusebi V, Ichihara S, Vincent-Salomon A et al, Exceptionally rare types and variants. In: Lakhani SR, Ellis IO, Schnitt SJ et al, (eds). WHO Classification of Tumours of the Breast, 4th edn. International Agency for Research on Cancer (IARC): Lyon, France, 2012, pp 71–76.
McDivitt RW, Stewart FW . Breast carcinoma in children. JAMA 1966;195:388–390.
Horowitz DP, Sharma CS, Connolly E et al, Secretory carcinoma of the breast: results from the survival, epidemiology and end results database. Breast 2012;21:350–353.
Tognon C, Knezevich SR, Huntsman D et al, Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2002;2:367–376.
Makretsov N, He M, Hayes M et al, A fluorescence in situ hybridization study of ETV6-NTRK3 fusion gene in secretory breast carcinoma. Genes Chromosomes Cancer 2004;40:152–157.
Jin W, Yun C, Hobbie A et al, Cellular transformation and activation of the phosphoinositide-3-kinase-Akt cascade by the ETV6-NTRK3 chimeric tyrosine kinase requires c-Src. Cancer Res 2007;67:3192–3200.
Lannon CL, Sorensen PH . ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin Cancer Biol 2005;15:215–223.
Morrison KB, Tognon CE, Garnett MJ et al, ETV6-NTRK3 transformation requires insulin-like growth factor 1 receptor signaling and is associated with constitutive IRS-1 tyrosine phosphorylation. Oncogene 2002;21:5684–5695.
Tognon C, Garnett M, Kenward E et al, The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res 2001;61:8909–8916.
Tognon CE, Martin MJ, Moradian A et al, A tripartite complex composed of ETV6-NTRK3, IRS1 and IGF1R is required for ETV6-NTRK3-mediated membrane localization and transformation. Oncogene 2012;31:1334–1340.
Wai DH, Knezevich SR, Lucas T et al, The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene 2000;19:906–915.
Lae M, Freneaux P, Sastre-Garau X et al, Secretory breast carcinomas with ETV6-NTRK3 fusion gene belong to the basal-like carcinoma spectrum. Mod Pathol 2009;22:291–298.
Li D, Xiao X, Yang W et al, Secretory breast carcinoma: a clinicopathological and immunophenotypic study of 15 cases with a review of the literature. Mod Pathol 2012;25:567–575.
Lee SG, Jung SP, Lee HY et al, Secretory breast carcinoma: a report of three cases and a review of the literature. Oncol Lett 2014;8:683–686.
Strauss BL, Bratthauer GL, Tavassoli FA . STAT 5a expression in the breast is maintained in secretory carcinoma, in contrast to other histologic types. Hum Pathol 2006;37:586–592.
Curtis C, Shah SP, Chin SF et al, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–352.
Dawson SJ, Rueda OM, Aparicio S et al, A new genome-driven integrated classification of breast cancer and its implications. EMBO J 2013;32:617–628.
Badve S, Dabbs DJ, Schnitt SJ et al, Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol 2011;24:157–167.
Banerji S, Cibulskis K, Rangel-Escareno C et al, Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012;486:405–409.
Nik-Zainal S, Davies H, Staaf J et al, Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54.
Stephens PJ, Tarpey PS, Davies H et al, The landscape of cancer genes and mutational processes in breast cancer. Nature 2012;486:400–404.
Pereira B, Chin SF, Rueda OM et al, The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479.
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70.
Shah SP, Roth A, Goya R et al, The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395–399.
Ciriello G, Gatza ML, Beck AH et al, Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–519.
Perou CM, Sorlie T, Eisen MB et al, Molecular portraits of human breast tumours. Nature 2000;406:747–752.
Lambros MB, Tan DS, Jones RL et al, Genomic profile of a secretory breast cancer with an ETV6-NTRK3 duplication. J Clin Pathol 2009;62:604–612.
Maitra A, Tavassoli FA, Albores-Saavedra J et al, Molecular abnormalities associated with secretory carcinomas of the breast. Hum Pathol 1999;30:1435–1440.
Amin SM, Beattie A, Ling X et al, Primary cutaneous mammary analog secretory carcinoma with ETV6-NTRK3 translocation. Am J Dermatopathol 2016;38:842–845.
Bishop JA . Unmasking MASC: bringing to light the unique morphologic, immunohistochemical and genetic features of the newly recognized mammary analogue secretory carcinoma of salivary glands. Head Neck Pathol 2013;7:35–39.
Bishop JA, Taube JM, Su A et al, Secretory carcinoma of the skin harboring ETV6 gene fusions: a cutaneous analogue to secretory carcinomas of the breast and salivary glands. Am J Surg Pathol 2017;41:62–66.
Chang MD, Arthur AK, Garcia JJ et al, ETV6 rearrangement in a case of mammary analogue secretory carcinoma of the skin. J Cutan Pathol 2016;43:1045–1049.
Connor A, Perez-Ordonez B, Shago M et al, Mammary analog secretory carcinoma of salivary gland origin with the ETV6 gene rearrangement by FISH: expanded morphologic and immunohistochemical spectrum of a recently described entity. Am J Surg Pathol 2012;36:27–34.
Dettloff J, Seethala RR, Stevens TM et al, Mammary analog secretory carcinoma (MASC) involving the thyroid gland: a report of the first 3 cases. Head Neck Pathol 2016; doi: 10.1007/s12105-016-0741-6; e-pub ahead of print.
Dogan S, Wang L, Ptashkin RN et al, Mammary analog secretory carcinoma of the thyroid gland: a primary thyroid adenocarcinoma harboring ETV6-NTRK3fusion. Mod Pathol 2016;29:985–995.
Majewska H, Skalova A, Stodulski D et al, Mammary analogue secretory carcinoma of salivary glands: a new entity associated with ETV6 gene rearrangement. Virchows Arch 2015;466:245–254.
Skalova A, Vanecek T, Sima R et al, Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol 2010;34:599–608.
Skalova A, Vanecek T, Simpson RH et al, Mammary analogue secretory carcinoma of salivary glands: molecular analysis of 25 ETV6 gene rearranged tumors with lack of detection of classical ETV6-NTRK3 fusion transcript by standard RT-PCR: report of 4 cases harboring ETV6-X gene fusion. Am J Surg Pathol 2016;40:3–13.
Leeman-Neill RJ, Kelly LM, Liu P et al, ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 2014;120:799–807.
Martelotto LG, De Filippo MR, Ng CKY et al, Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol 2015;237:179–189.
Persson M, Andren Y, Mark J et al, Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA 2009;106:18740–18744.
Ghabach B, Anderson WF, Curtis RE et al, Adenoid cystic carcinoma of the breast in the United States (1977 to 2006): a population-based cohort study. Breast Cancer Res 2010;12:R54.
Pia-Foschini M, Reis-Filho JS, Eusebi V et al, Salivary gland-like tumours of the breast: surgical and molecular pathology. J Clin Pathol 2003;56:497–506.
Camelo-Piragua SI, Habib C, Kanumuri P et al, Mucoepidermoid carcinoma of the breast shares cytogenetic abnormality with mucoepidermoid carcinoma of the salivary gland: a case report with molecular analysis and review of the literature. Hum Pathol 2009;40:887–892.
Di Tommaso L, Foschini MP, Ragazzini T et al, Mucoepidermoid carcinoma of the breast. Virchows Arch 2004;444:13–19.
Fisher ER, Palekar AS, Gregorio RM et al, Mucoepidermoid and squamous cell carcinomas of breast with reference to squamous metaplasia and giant cell tumors. Am J Surg Pathol 1983;7:15–27.
Guerini-Rocco E, Hodi Z, Piscuoglio S et al, The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol 2015;237:166–178.
Piscuoglio S, Hodi Z, Katabi N et al, Are acinic cell carcinomas of the breast and salivary glands distinct diseases? Histopathology 2015;67:529–537.
Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM.: Broad Institute. Available from http://broadinstitute.github.io/picard.
Garrison E, Marth G . Haplotype-based variant detection from short-read sequencing arXiv 2012;1207.3907 [q-bio.GN].
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–595.
Li H, Handsaker B, Wysoker A et al, The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079.
McKenna A, Hanna M, Banks E et al, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303.
Rausch T, Zichner T, Schlattl A et al, DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333–i339.
Talevich E, Shain AH, Botton T et al, CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873.
Van der Auwera GA, Carneiro MO, Hartl C et al, From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11 0 1–11 033.
Yang H, Wang K . Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 2015;10:1556–1566.
Ye K, Schulz MH, Long Q et al, Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865–2871.
DePristo MA, Banks E, Poplin R et al, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–498.
Krings G, Nystrom M, Mehdi I et al, Diagnostic utility and sensitivities of GATA3 antibodies in triple-negative breast cancer. Hum Pathol 2014;45:2225–2232.
Hammond ME, Hayes DF, Dowsett M et al, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med 2010;134:e48–e72.
Wolff AC, Hammond ME, Hicks DG et al, Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013.
Lopez-Garcia MA, Geyer FC, Lacroix-Triki M et al, Breast cancer precursors revisited: molecular features and progression pathways. Histopathology 2010;57:171–192.
Alassiri AH, Ali RH, Shen Y et al, ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am J Surg Pathol 2016;40:1051–1061.
Arce C, Cortes-Padilla D, Huntsman DG et al, Secretory carcinoma of the breast containing the ETV6-NTRK3 fusion gene in a male: case report and review of the literature. World J Surg Oncol 2005;3:35.
Bourgeois JM, Knezevich SR, Mathers JA et al, Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol 2000;24:937–946.
El Demellawy D, Cundiff CA, Nasr A et al, Congenital mesoblastic nephroma: a study of 19 cases using immunohistochemistry and ETV6-NTRK3 fusion gene rearrangement. Pathology 2016;48:47–50.
Fehr A, Loning T, Stenman G . Mammary analogue secretory carcinoma of the salivary glands with ETV6-NTRK3 gene fusion. Am J Surg Pathol 2011;35:1600–1602.
Knezevich SR, McFadden DE, Tao W et al, A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998;18:184–187.
Kralik JM, Kranewitter W, Boesmueller H et al, Characterization of a newly identified ETV6-NTRK3 fusion transcript in acute myeloid leukemia. Diagn Pathol 2011;6:19.
Rubin BP, Chen CJ, Morgan TW et al, Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol 1998;153:1451–1458.
Cetinbas N, Huang-Hobbs H, Tognon C et al, Mutation of the salt bridge-forming residues in the ETV6-SAM domain interface blocks ETV6-NTRK3-induced cellular transformation. J Biol Chem 2013;288:27940–27950.
Tognon CE, Mackereth CD, Somasiri AM et al, Mutations in the SAM domain of the ETV6-NTRK3 chimeric tyrosine kinase block polymerization and transformation activity. Mol Cell Biol 2004;24:4636–4650.
Ito Y, Ishibashi K, Masaki A et al, Mammary analogue secretory carcinoma of salivary glands: a clinicopathologic and molecular study including 2 cases harboring ETV6-X fusion. Am J Surg Pathol 2015;39:602–610.
Choo JR, Nielsen TO . Biomarkers for basal-like breast cancer. Cancers (Basel) 2010;2:1040–1065.
Ellis IO . Intraductal proliferative lesions of the breast: morphology, associated risk and molecular biology. Mod Pathol 2010;23 (Suppl 2):S1–S7.
Said-Al-Naief N, Carlos R, Vance GH et al. Combined DOG1 and mammaglobin immunohistochemistry is comparable to ETV6-breakapart analysis for differentiating between papillary cystic variants of acinic cell carcinoma and mammary analogue secretory carcinoma. Int J Surg Pathol 2017;25:127–140.
Bishop JA, Yonescu R, Batista D et al, Utility of mammaglobin immunohistochemistry as a proxy marker for the ETV6-NTRK3 translocation in the diagnosis of salivary mammary analogue secretory carcinoma. Hum Pathol 2013;44:1982–1988.
Hsieh MS, Lee YH, Chang YL . SOX10-positive salivary gland tumors: a growing list, including mammary analogue secretory carcinoma of the salivary gland, sialoblastoma, low-grade salivary duct carcinoma, basal cell adenoma/adenocarcinoma, and a subgroup of mucoepidermoid carcinoma. Hum Pathol 2016;56:134–142.
Acknowledgements
We thank the referring pathologists for providing the cases included in this study. We also thank Jie Hou for technical expertise in fluorescence in situ hybridization studies. This study was funded by the Department of Pathology of the University of California, San Francisco.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Krings, G., Joseph, N., Bean, G. et al. Genomic profiling of breast secretory carcinomas reveals distinct genetics from other breast cancers and similarity to mammary analog secretory carcinomas. Mod Pathol 30, 1086–1099 (2017). https://doi.org/10.1038/modpathol.2017.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.32
This article is cited by
-
Clinicopathologic and genetic analysis of invasive breast carcinomas in women with germline CHEK2 variants
Breast Cancer Research and Treatment (2024)
-
Clinicopathological investigation of secretory carcinoma cases including a successful treatment outcome using entrectinib for high-grade transformation: a case report
BMC Medical Genomics (2022)
-
Clinicopathological features and genomic profiles of a group of secretory breast carcinomas in which progressive cases have more complex genomic features
Diagnostic Pathology (2022)
-
PTHrP induces STAT5 activation, secretory differentiation and accelerates mammary tumor development
Breast Cancer Research (2022)
-
AKT-mediated phosphorylation of Sox9 induces Sox10 transcription in a murine model of HER2-positive breast cancer
Breast Cancer Research (2021)